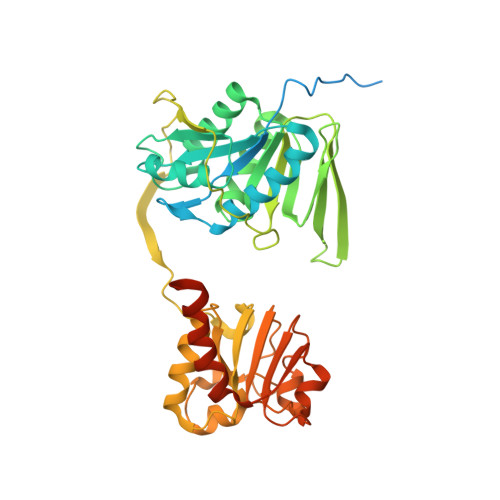
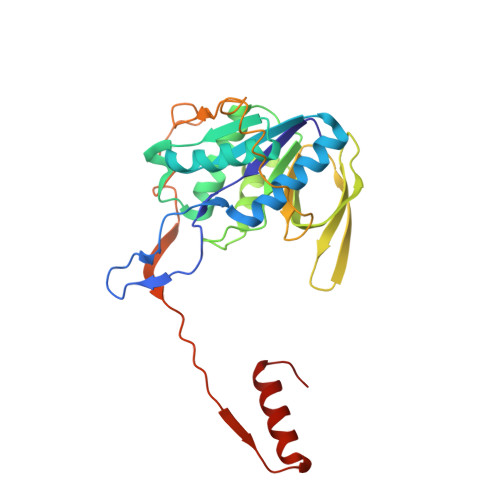
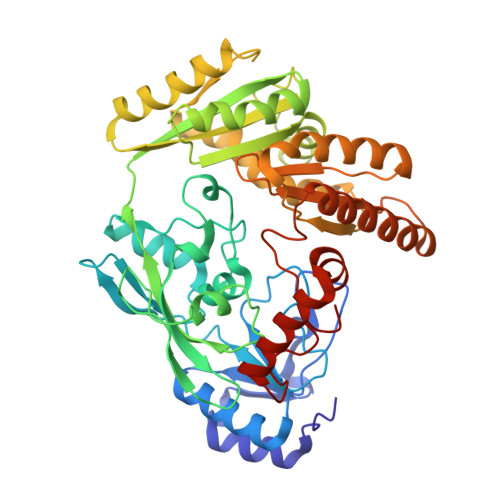
Structure-based electron-confurcation mechanism of the Ldh-EtfAB complex.
Kayastha, K., Katsyv, A., Himmrich, C., Welsch, S., Schuller, J.M., Ermler, U., Muller, V.(2022) Elife 11
- PubMed: 35748623
- DOI: https://doi.org/10.7554/eLife.77095
- Primary Citation of Related Structures:
7QH2 - PubMed Abstract:
Lactate oxidation with NAD + as electron acceptor is a highly endergonic reaction. Some anaerobic bacteria overcome the energetic hurdle by flavin-based electron bifurcation/confurcation (FBEB/FBEC) using a lactate dehydrogenase (Ldh) in concert with the electron-transferring proteins EtfA and EtfB. The electron cryo-microscopically characterized (Ldh-EtfAB) 2 complex of Acetobacterium woodii at 2.43 Å resolution consists of a mobile EtfAB shuttle domain located between the rigid central Ldh and the peripheral EtfAB base units. The FADs of Ldh and the EtfAB shuttle domain contact each other thereby forming the D (dehydrogenation-connected) state. The intermediary Glu37 and Glu139 may harmonize the redox potentials between the FADs and the pyruvate/lactate pair crucial for FBEC. By integrating Alphafold2 calculations a plausible novel B (bifurcation-connected) state was obtained allowing electron transfer between the EtfAB base and shuttle FADs. Kinetic analysis of enzyme variants suggests a correlation between NAD + binding site and D-to-B-state transition implicating a 75° rotation of the EtfAB shuttle domain. The FBEC inactivity when truncating the ferredoxin domain of EtfA substantiates its role as redox relay. Lactate oxidation in Ldh is assisted by the catalytic base His423 and a metal center. On this basis, a comprehensive catalytic mechanism of the FBEC process was proposed.
- Departments of Molecular Membrane Biology of the Max-Planck-Institut for Biophysics, Frankfurt am Main, Germany.
Organizational Affiliation: