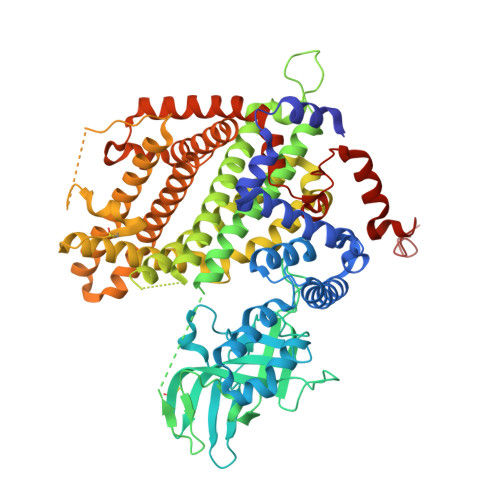
Crystal structures of apo- and FAD-bound human peroxisomal acyl-CoA oxidase provide mechanistic basis explaining clinical observations.
Sonani, R.R., Blat, A., Dubin, G.(2022) Int J Biol Macromol 205: 203-210
- PubMed: 35149097
- DOI: https://doi.org/10.1016/j.ijbiomac.2022.02.008
- Primary Citation of Related Structures:
7Q84, 7Q86 - PubMed Abstract:
Peroxisomal acyl-CoA oxidase 1a (ACOX1a) catalyzes the first and rate-limiting step of fatty acid oxidation, the conversion of acyl-CoAs to 2-trans-enoyl-CoAs. The dysfunction of human ACOX1a (hACOX1a) leads to deterioration of the nervous system manifesting in myeloneuropathy, hypotonia and convulsions. Crystal structures of hACOX1a in apo- and cofactor (FAD)-bound forms were solved at 2.00 and 2.09 Å resolution, respectively. hACOX1a exists as a homo-dimer with solvation free energy gain (ΔG o ) of -44.7 kcal mol -1 . Two FAD molecules bind at the interface of protein monomers completing the active sites. The substrate binding cleft of hACOX1a is wider compared to human mitochondrial very-long chain specific acyl-CoA dehydrogenase. Mutations (p.G178C, p.M278V and p.N237S) reported to cause dysfunctionality of hACOX1a are analyzed on its 3D-structure to understand structure-function related perturbations and explain the associated phenotypes.
- Malopolska Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, Poland.
Organizational Affiliation: