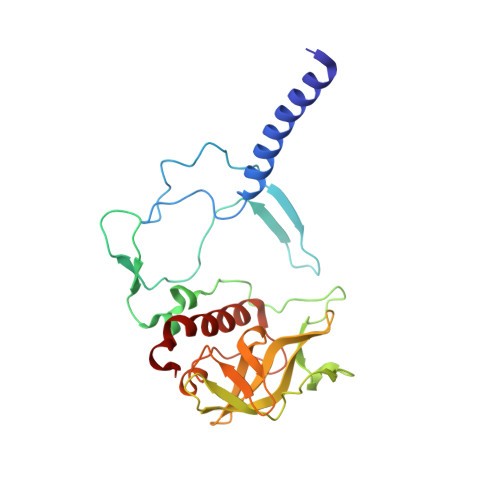
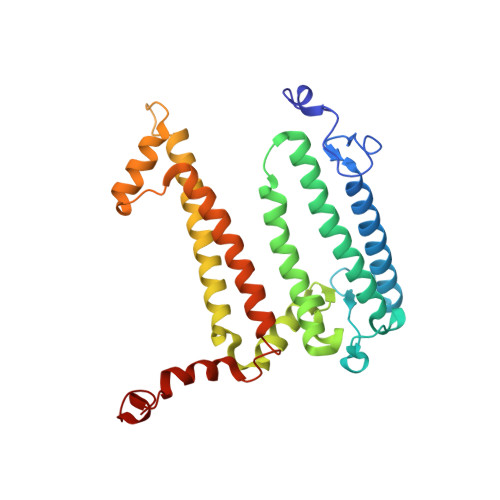
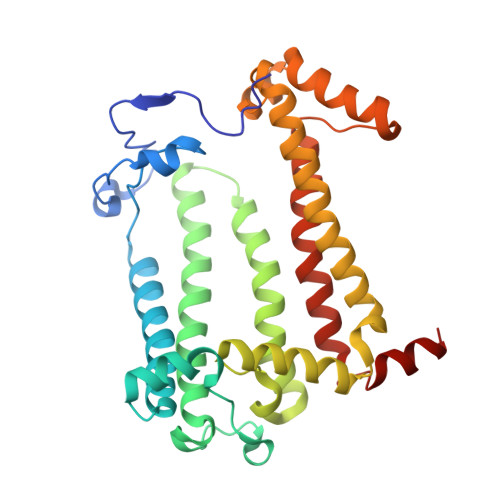
Fixed-target high-pressure macromolecular crystallography
Lieske, J., Saouane, S., Guenther, S., Meyer, J., Pakendorf, T., Reime, B., Burkhardt, A., Crosas, E., Hakanpaeae, J., Stachnik, K., Sieg, J., Rarey, M., Abdellatif, M.H., Gabdulkhakov, A.G., Selikhanov, G.K., Chapman, H.N., Meents, A.To be published.