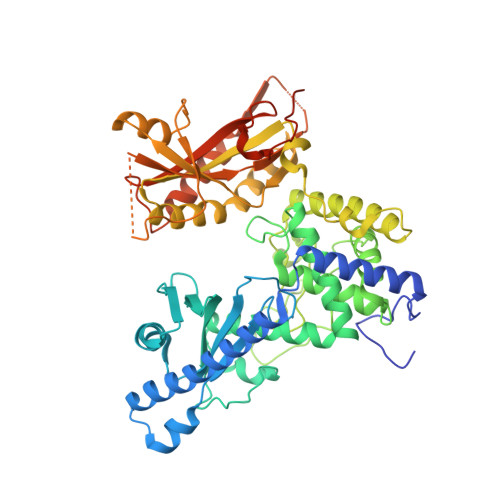
Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Soni, K., Sivadas, A., Horvath, A., Dobrev, N., Hayashi, R., Kiss, L., Simon, B., Wild, K., Sinning, I., Fischer, T.(2023) Nat Commun 14: 772-772
- PubMed: 36774373
- DOI: https://doi.org/10.1038/s41467-023-36402-6
- Primary Citation of Related Structures:
7Q72, 7Q73, 7Q74 - PubMed Abstract:
The S. pombe orthologue of the human PAXT connection, Mtl1-Red1 Core (MTREC), is an eleven-subunit complex that targets cryptic unstable transcripts (CUTs) to the nuclear RNA exosome for degradation. It encompasses the canonical poly(A) polymerase Pla1, responsible for polyadenylation of nascent RNA transcripts as part of the cleavage and polyadenylation factor (CPF/CPSF). In this study we identify and characterise the interaction between Pla1 and the MTREC complex core component Red1 and analyse the functional relevance of this interaction in vivo. Our crystal structure of the Pla1-Red1 complex shows that a 58-residue fragment in Red1 binds to the RNA recognition motif domain of Pla1 and tethers it to the MTREC complex. Structure-based Pla1-Red1 interaction mutations show that Pla1, as part of MTREC complex, hyper-adenylates CUTs for their efficient degradation. Interestingly, the Red1-Pla1 interaction is also required for the efficient assembly of the fission yeast facultative heterochromatic islands. Together, our data suggest a complex interplay between the RNA surveillance and 3'-end processing machineries.
- Heidelberg University Biochemistry Center (BZH), INF 328, D-69120, Heidelberg, Germany.
Organizational Affiliation: