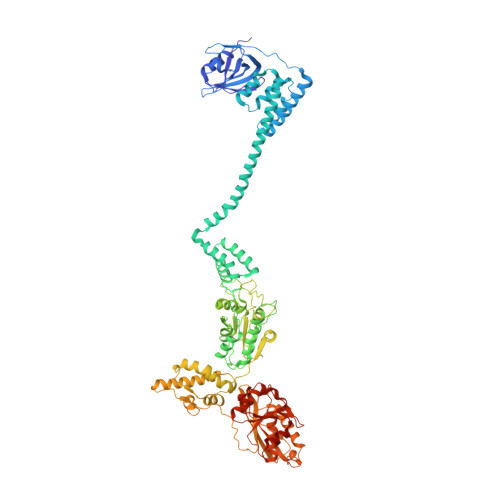
Cryo-EM structure of the full-length Lon protease from Thermus thermophilus.
Coscia, F., Lowe, J.(2021) FEBS Lett 595: 2691-2700
- PubMed: 34591981
- DOI: https://doi.org/10.1002/1873-3468.14199
- Primary Citation of Related Structures:
7P6U - PubMed Abstract:
In bacteria, Lon is a large hexameric ATP-dependent protease that targets misfolded and also folded substrates, some of which are involved in cell division and survival of cellular stress. The N-terminal domain of Lon facilitates substrate recognition, but how the domains confer such activity has remained unclear. Here, we report the full-length structure of Lon protease from Thermus thermophilus at 3.9 Å resolution in a substrate-engaged state. The six N-terminal domains are arranged in three pairs, stabilized by coiled-coil segments and forming an additional channel for substrate sensing and entry into the AAA+ ring. Sequence conservation analysis and proteolysis assays confirm that this architecture is required for the degradation of both folded and unfolded substrates in bacteria.
- MRC Laboratory of Molecular Biology, Cambridge Biomedical Campus, Cambridge, UK.
Organizational Affiliation: