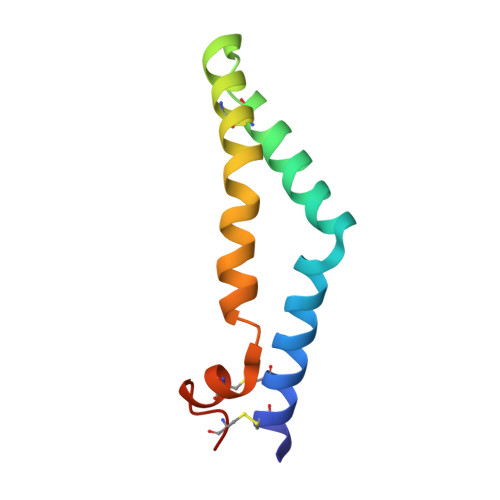
A Tetrameric Assembly of Saposin A: Increasing Structural Diversity in Lipid Transfer Proteins.
Shamin, M., Spratley, S.J., Graham, S.C., Deane, J.E.(2021) Contact (Thousand Oaks) 4: 251525642110523-251525642110523
- PubMed: 37143956
- DOI: https://doi.org/10.1177/25152564211052382
- Primary Citation of Related Structures:
7P4T - PubMed Abstract:
Saposins are lipid transfer proteins required for the degradation of sphingolipids in the lysosome. These small proteins bind lipids by transitioning from a closed, monomeric state to an open conformation exposing a hydrophobic surface that binds and shields hydrophobic lipid tails from the aqueous environment. Saposins form a range of multimeric assemblies to encompass these bound lipids and present them to hydrolases in the lysosome. This lipid-binding property of human saposin A has been exploited to form lipoprotein nanodiscs suitable for structural studies of membrane proteins. Here we present the crystal structure of a unique tetrameric assembly of murine saposin A produced serendipitously, following modifications of published protocols for making lipoprotein nanodiscs. The structure of this new saposin oligomer highlights the diversity of tertiary arrangement that can be adopted by these important lipid transfer proteins.
- Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 0XY, UK.
Organizational Affiliation: