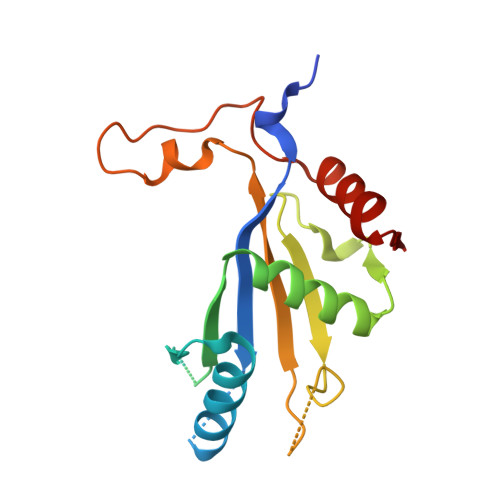
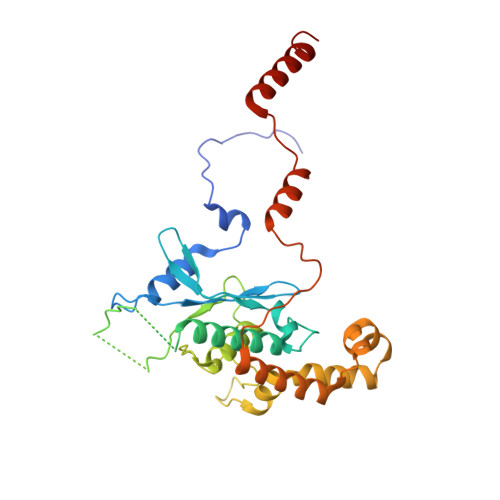
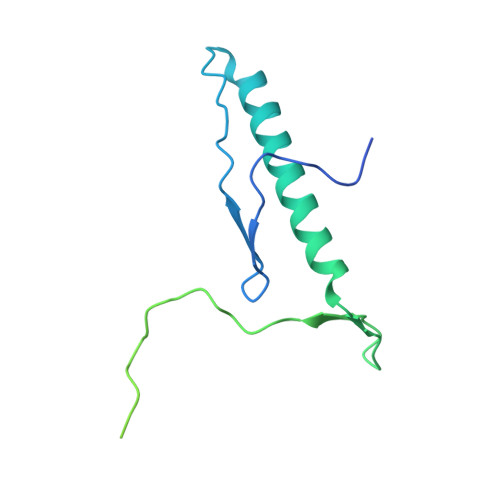
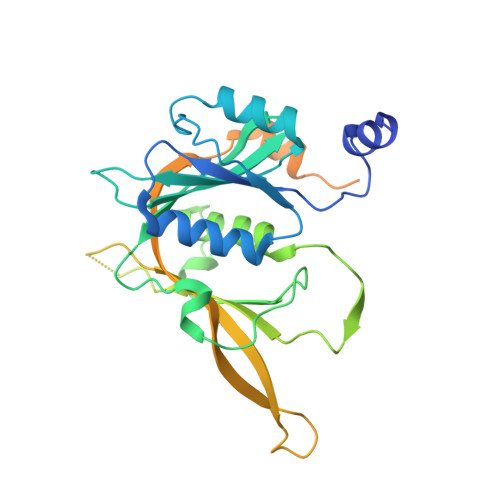
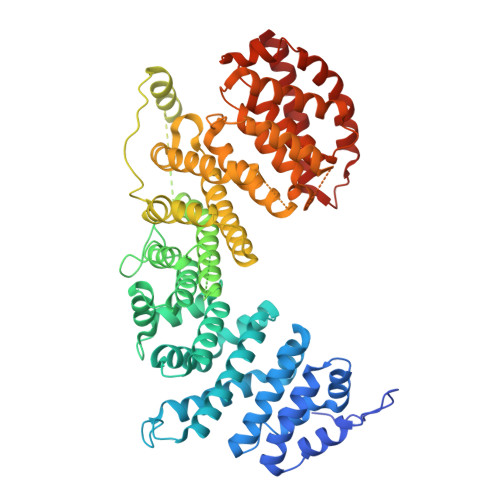
Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Castillo Duque de Estrada, N.M., Thoms, M., Flemming, D., Hammaren, H.M., Buschauer, R., Ameismeier, M., Bassler, J., Beck, M., Beckmann, R., Hurt, E.(2023) Nat Struct Mol Biol
- PubMed: 37291423
- DOI: https://doi.org/10.1038/s41594-023-01006-7
- Primary Citation of Related Structures:
7OZS, 8BGU - PubMed Abstract:
The 5S ribonucleoprotein (RNP) is assembled from its three components (5S rRNA, Rpl5/uL18 and Rpl11/uL5) before being incorporated into the pre-60S subunit. However, when ribosome synthesis is disturbed, a free 5S RNP can enter the MDM2-p53 pathway to regulate cell cycle and apoptotic signaling. Here we reconstitute and determine the cryo-electron microscopy structure of the conserved hexameric 5S RNP with fungal or human factors. This reveals how the nascent 5S rRNA associates with the initial nuclear import complex Syo1-uL18-uL5 and, upon further recruitment of the nucleolar factors Rpf2 and Rrs1, develops into the 5S RNP precursor that can assemble into the pre-ribosome. In addition, we elucidate the structure of another 5S RNP intermediate, carrying the human ubiquitin ligase Mdm2, which unravels how this enzyme can be sequestered from its target substrate p53. Our data provide molecular insight into how the 5S RNP can mediate between ribosome biogenesis and cell proliferation.
- Heidelberg University Biochemistry Center (BZH), Heidelberg, Germany.
Organizational Affiliation: