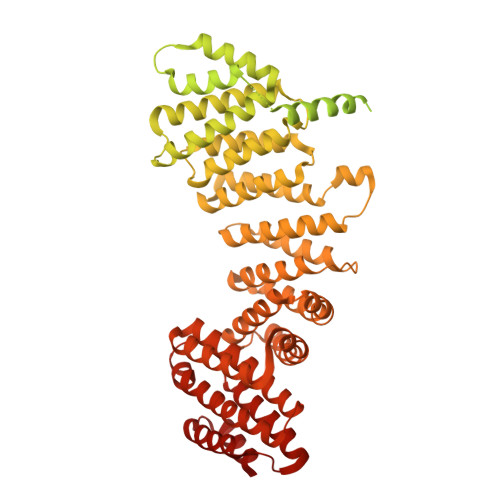
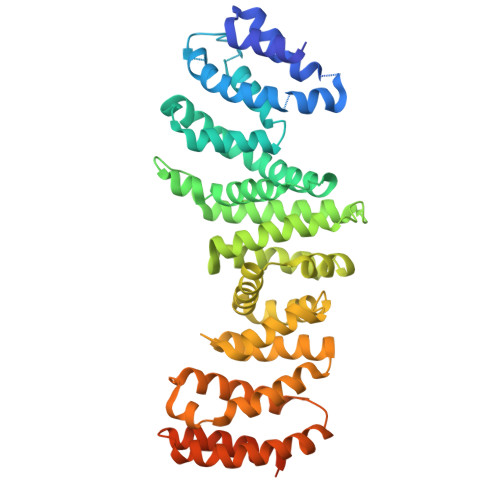
Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Pal, M., Munoz-Hernandez, H., Bjorklund, D., Zhou, L., Degliesposti, G., Skehel, J.M., Hesketh, E.L., Thompson, R.F., Pearl, L.H., Llorca, O., Prodromou, C.(2021) Cell Rep 36: 109317-109317
- PubMed: 34233195
- DOI: https://doi.org/10.1016/j.celrep.2021.109317
- Primary Citation of Related Structures:
7OLE - PubMed Abstract:
The R2TP (RUVBL1-RUVBL2-RPAP3-PIH1D1) complex, in collaboration with heat shock protein 90 (HSP90), functions as a chaperone for the assembly and stability of protein complexes, including RNA polymerases, small nuclear ribonucleoprotein particles (snRNPs), and phosphatidylinositol 3-kinase (PI3K)-like kinases (PIKKs) such as TOR and SMG1. PIKK stabilization depends on an additional complex of TELO2, TTI1, and TTI2 (TTT), whose structure and function are poorly understood. The cryoelectron microscopy (cryo-EM) structure of the human R2TP-TTT complex, together with biochemical experiments, reveals the mechanism of TOR recruitment to the R2TP-TTT chaperone. The HEAT-repeat TTT complex binds the kinase domain of TOR, without blocking its activity, and delivers TOR to the R2TP chaperone. In addition, TTT regulates the R2TP chaperone by inhibiting RUVBL1-RUVBL2 ATPase activity and by modulating the conformation and interactions of the PIH1D1 and RPAP3 components of R2TP. Taken together, our results show how TTT couples the recruitment of TOR to R2TP with the regulation of this chaperone system.
- Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Falmer, Brighton BN1 9RQ, UK.
Organizational Affiliation: