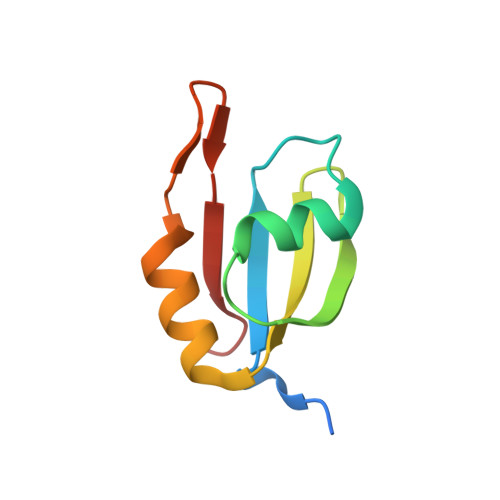
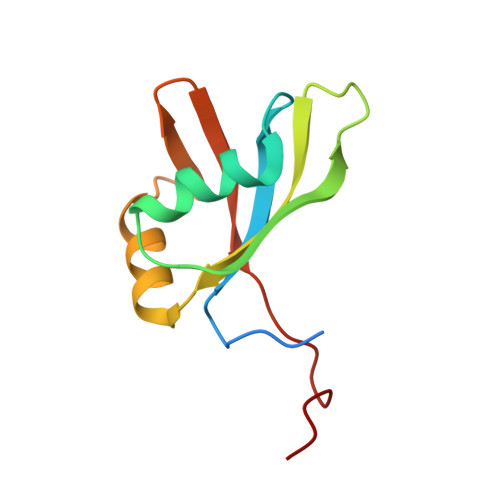
Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Perez-Borrajero, C., Podvalnaya, N., Holleis, K., Lichtenberger, R., Karaulanov, E., Simon, B., Basquin, J., Hennig, J., Ketting, R.F., Falk, S.(2021) Genes Dev 35: 1304-1323
- PubMed: 34413138
- DOI: https://doi.org/10.1101/gad.348648.121
- Primary Citation of Related Structures:
7O6L, 7O6N, 7OCX, 7OCZ - PubMed Abstract:
Piwi-interacting RNAs (piRNAs) constitute a class of small RNAs that bind PIWI proteins and are essential to repress transposable elements in the animal germline, thereby promoting genome stability and maintaining fertility. C. elegans piRNAs (21U RNAs) are transcribed individually from minigenes as precursors that require 5' and 3' processing. This process depends on the PETISCO complex, consisting of four proteins: IFE-3, TOFU-6, PID-3, and ERH-2. We used biochemical and structural biology approaches to characterize the PETISCO architecture and its interaction with RNA, together with its effector proteins TOST-1 and PID-1. These two proteins define different PETISCO functions: PID-1 governs 21U processing, whereas TOST-1 links PETISCO to an unknown process essential for early embryogenesis. Here, we show that PETISCO forms an octameric assembly with each subunit present in two copies. Determination of structures of the TOFU-6/PID-3 and PID-3/ERH-2 subcomplexes, supported by in vivo studies of subunit interaction mutants, allows us to propose a model for the formation of the TOFU-6/PID-3/ERH-2 core complex and its functionality in germ cells and early embryos. Using NMR spectroscopy, we demonstrate that TOST-1 and PID-1 bind to a common surface on ERH-2, located opposite its PID-3 binding site, explaining how PETISCO can mediate different cellular roles.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL) Heidelberg, 69117 Heidelberg, Germany.
Organizational Affiliation: