Discovery and Characterization of Potent Dual P-Glycoprotein and CYP3A4 Inhibitors: Design, Synthesis, Cryo-EM Analysis, and Biological Evaluations.
Urgaonkar, S., Nosol, K., Said, A.M., Nasief, N.N., Bu, Y., Locher, K.P., Lau, J.Y.N., Smolinski, M.P.(2022) J Med Chem 65: 191-216
- PubMed: 34928144
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01272
- Primary Citation of Related Structures:
7O9W - PubMed Abstract:
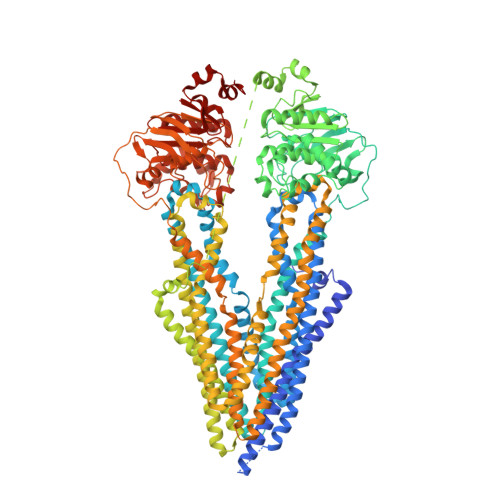
Targeted concurrent inhibition of intestinal drug efflux transporter P-glycoprotein (P-gp) and drug metabolizing enzyme cytochrome P450 3A4 (CYP3A4) is a promising approach to improve oral bioavailability of their common substrates such as docetaxel, while avoiding side effects arising from their pan inhibitions. Herein, we report the discovery and characterization of potent small molecule inhibitors of P-gp and CYP3A4 with encequidar (minimally absorbed P-gp inhibitor) as a starting point for optimization. To aid in the design of these dual inhibitors, we solved the high-resolution cryo-EM structure of encequidar bound to human P-gp. The structure guided us to prudently decorate the encequidar scaffold with CYP3A4 pharmacophores, leading to the identification of several analogues with dual potency against P-gp and CYP3A4. In vivo , dual P-gp and CYP3A4 inhibitor 3a improved the oral absorption of docetaxel by 3-fold as compared to vehicle, while 3a itself remained poorly absorbed.
- Athenex Inc., Conventus Building, Buffalo, New York 14203, United States.
Organizational Affiliation: