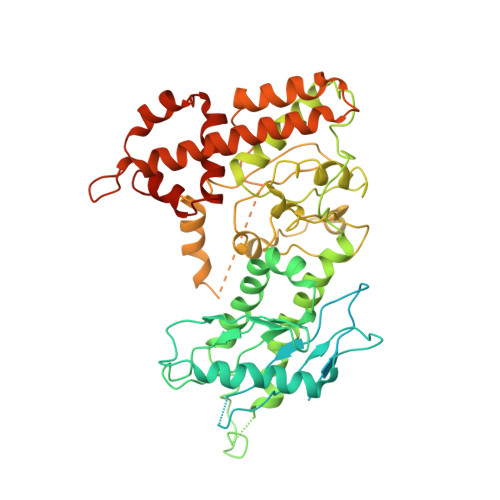
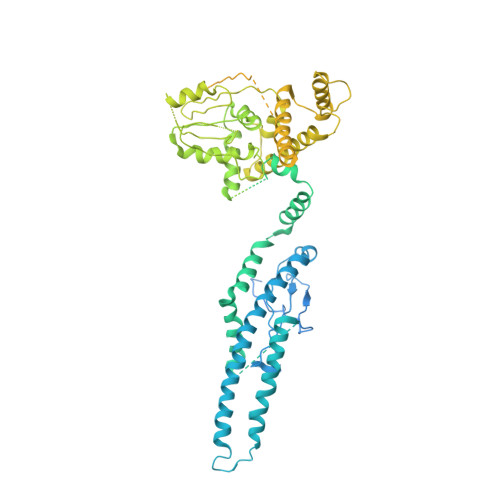
Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.
Norpel, J., Cavadini, S., Schenk, A.D., Graff-Meyer, A., Hess, D., Seebacher, J., Chao, J.A., Bhaskar, V.(2021) PLoS Biol 19: e3001344-e3001344
- PubMed: 34297726
- DOI: https://doi.org/10.1371/journal.pbio.3001344
- Primary Citation of Related Structures:
7O2W - PubMed Abstract:
A major cause of familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) spectrum disorder is the hexanucleotide G4C2 repeat expansion in the first intron of the C9orf72 gene. Many underlying mechanisms lead to manifestation of disease that include toxic gain-of-function by repeat G4C2 RNAs, dipeptide repeat proteins, and a reduction of the C9orf72 gene product. The C9orf72 protein interacts with SMCR8 and WDR41 to form a trimeric complex and regulates multiple cellular pathways including autophagy. Here, we report the structure of the C9orf72-SMCR8 complex at 3.8 Å resolution using single-particle cryo-electron microscopy (cryo-EM). The structure reveals 2 distinct dimerization interfaces between C9orf72 and SMCR8 that involves an extensive network of interactions. Homology between C9orf72-SMCR8 and Folliculin-Folliculin Interacting Protein 2 (FLCN-FNIP2), a GTPase activating protein (GAP) complex, enabled identification of a key residue within the active site of SMCR8. Further structural analysis suggested that a coiled-coil region within the uDenn domain of SMCR8 could act as an interaction platform for other coiled-coil proteins, and its deletion reduced the interaction of the C9orf72-SMCR8 complex with FIP200 upon starvation. In summary, this study contributes toward our understanding of the biological function of the C9orf72-SMCR8 complex.
- Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: