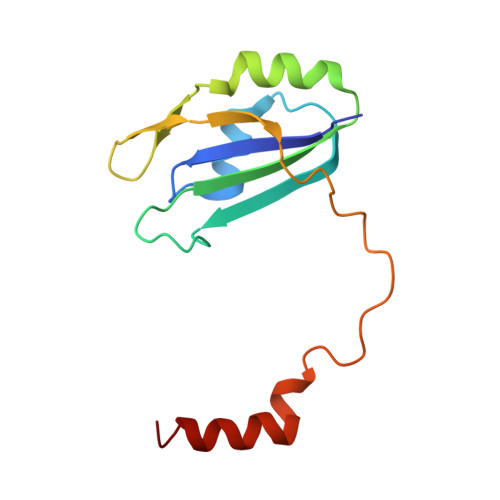
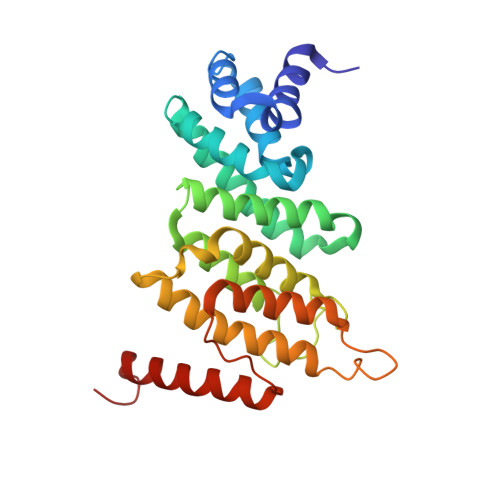
Structures of nonsense-mediated mRNA decay factors UPF3B and UPF3A in complex with UPF2 reveal molecular basis for competitive binding and for neurodevelopmental disorder-causing mutation.
Bufton, J.C., Powers, K.T., Szeto, J.A., Toelzer, C., Berger, I., Schaffitzel, C.(2022) Nucleic Acids Res 50: 5934-5947
- PubMed: 35640974
- DOI: https://doi.org/10.1093/nar/gkac421
- Primary Citation of Related Structures:
7NWU, 7QG6 - PubMed Abstract:
UPF3 is a key nonsense-mediated mRNA decay (NMD) factor required for mRNA surveillance and eukaryotic gene expression regulation. UPF3 exists as two paralogs (A and B) which are differentially expressed depending on cell type and developmental stage and believed to regulate NMD activity based on cellular requirements. UPF3B mutations cause intellectual disability. The underlying molecular mechanisms remain elusive, as many of the mutations lie in the poorly characterized middle-domain of UPF3B. Here, we show that UPF3A and UPF3B share structural and functional homology to paraspeckle proteins comprising an RNA-recognition motif-like domain (RRM-L), a NONA/paraspeckle-like domain (NOPS-L), and extended α-helical domain. These domains are essential for RNA/ribosome-binding, RNA-induced oligomerization and UPF2 interaction. Structures of UPF2's third middle-domain of eukaryotic initiation factor 4G (MIF4GIII) in complex with either UPF3B or UPF3A reveal unexpectedly intimate binding interfaces. UPF3B's disease-causing mutation Y160D in the NOPS-L domain displaces Y160 from a hydrophobic cleft in UPF2 reducing the binding affinity ∼40-fold compared to wildtype. UPF3A, which is upregulated in patients with the UPF3B-Y160D mutation, binds UPF2 with ∼10-fold higher affinity than UPF3B reliant mainly on NOPS-L residues. Our characterization of RNA- and UPF2-binding by UPF3's middle-domain elucidates its essential role in NMD.
- School of Biochemistry, University of Bristol; University Walk, Bristol BS8 1TD, UK.
Organizational Affiliation: