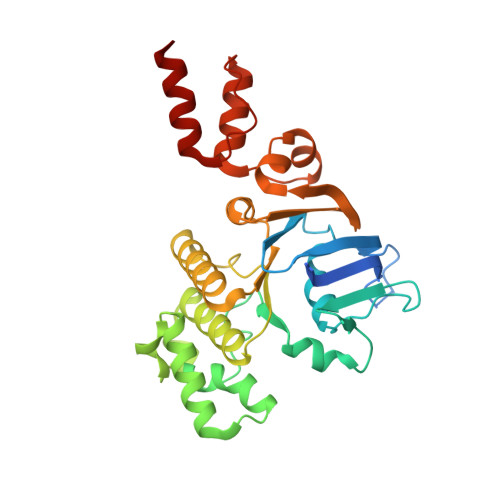
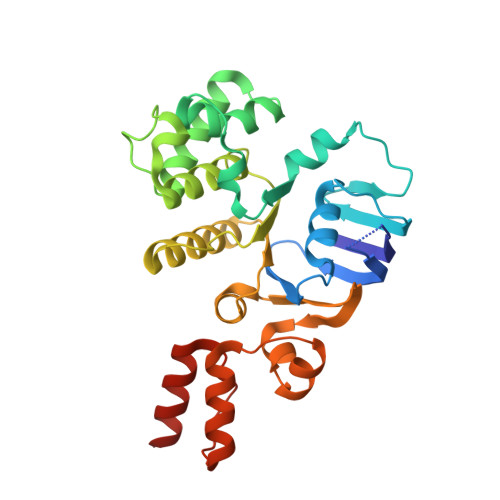
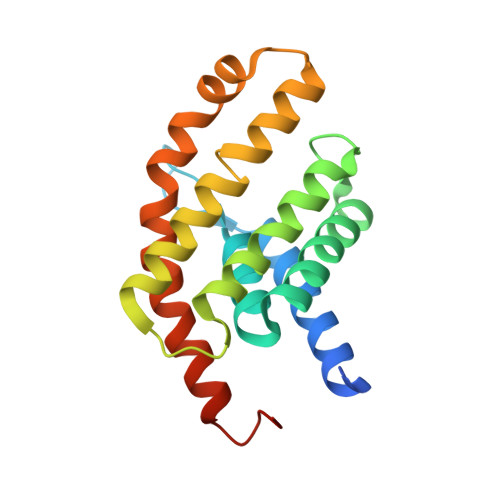
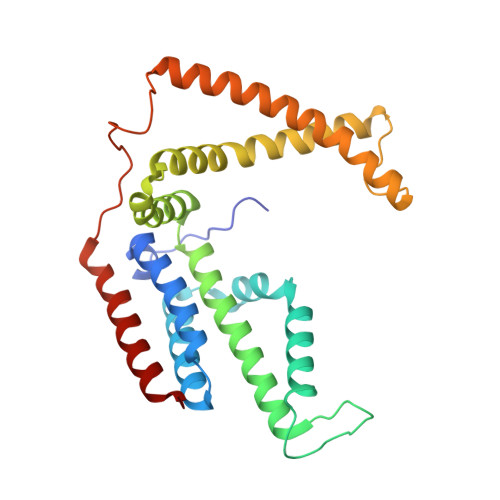
Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM.
Thangaratnarajah, C., Rheinberger, J., Paulino, C., Slotboom, D.J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34408021
- DOI: https://doi.org/10.1073/pnas.2105014118
- Primary Citation of Related Structures:
7NNT, 7NNU - PubMed Abstract:
Energy-coupling factor (ECF)-type transporters are small, asymmetric membrane protein complexes (∼115 kDa) that consist of a membrane-embedded, substrate-binding protein (S component) and a tripartite ATP-hydrolyzing module (ECF module). They import micronutrients into bacterial cells and have been proposed to use a highly unusual transport mechanism, in which the substrate is dragged across the membrane by a toppling motion of the S component. However, it remains unclear how the lipid bilayer could accommodate such a movement. Here, we used cryogenic electron microscopy at 200 kV to determine structures of a folate-specific ECF transporter in lipid nanodiscs and detergent micelles at 2.7- and 3.4-Å resolution, respectively. The structures reveal an irregularly shaped bilayer environment around the membrane-embedded complex and suggest that toppling of the S component is facilitated by protein-induced membrane deformations. In this way, structural remodeling of the lipid bilayer environment is exploited to guide the transport process.
- Faculty of Science and Engineering, Groningen Biomolecular Sciences and Biotechnology, Membrane Enzymology Group, University of Groningen, 9747 AG, Groningen, Netherlands.
Organizational Affiliation: