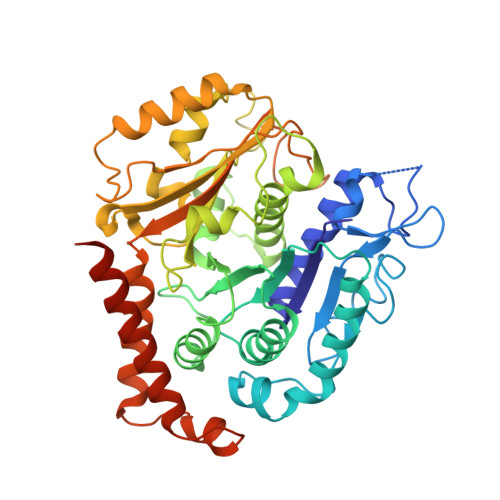
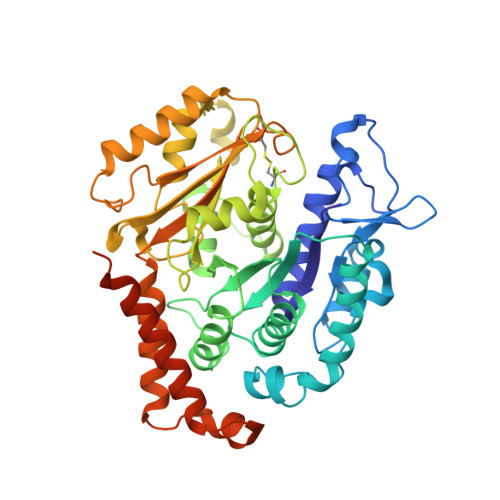
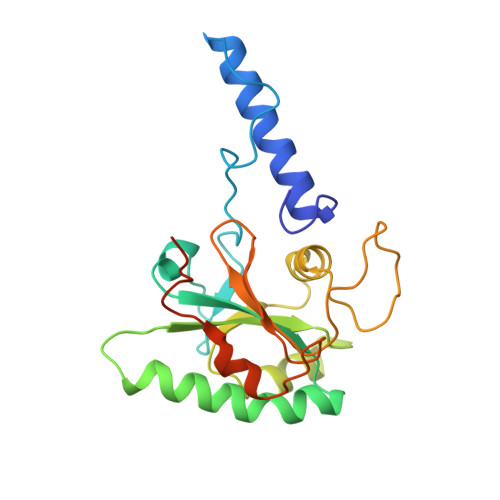
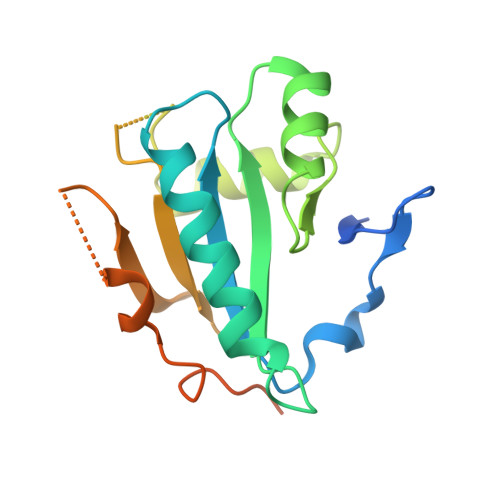
Cryo-EM structure of cortical microtubules from human parasite Toxoplasma gondii identifies their microtubule inner proteins.
Wang, X., Fu, Y., Beatty, W.L., Ma, M., Brown, A., David Sibley, L., Zhang, R.(2021) Nat Commun 12: 3065-3065
- PubMed: 34031406
- DOI: https://doi.org/10.1038/s41467-021-23351-1
- Primary Citation of Related Structures:
7MIZ - PubMed Abstract:
In living cells, microtubules (MTs) play pleiotropic roles, which require very different mechanical properties. Unlike the dynamic MTs found in the cytoplasm of metazoan cells, the specialized cortical MTs from Toxoplasma gondii, a prevalent human pathogen, are extraordinarily stable and resistant to detergent and cold treatments. Using single-particle cryo-EM, we determine their ex vivo structure and identify three proteins (TrxL1, TrxL2 and SPM1) as bona fide microtubule inner proteins (MIPs). These three MIPs form a mesh on the luminal surface and simultaneously stabilize the tubulin lattice in both longitudinal and lateral directions. Consistent with previous observations, deletion of the identified MIPs compromises MT stability and integrity under challenges by chemical treatments. We also visualize a small molecule like density at the Taxol-binding site of β-tubulin. Our results provide the structural basis to understand the stability of cortical MTs and suggest an evolutionarily conserved mechanism of MT stabilization from the inside.
- Department of Biochemistry and Molecular Biophysics, Washington University in St. Louis, School of Medicine, St. Louis, MO, USA.
Organizational Affiliation: