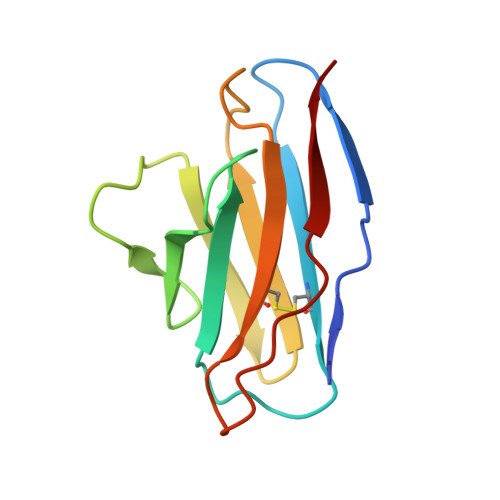
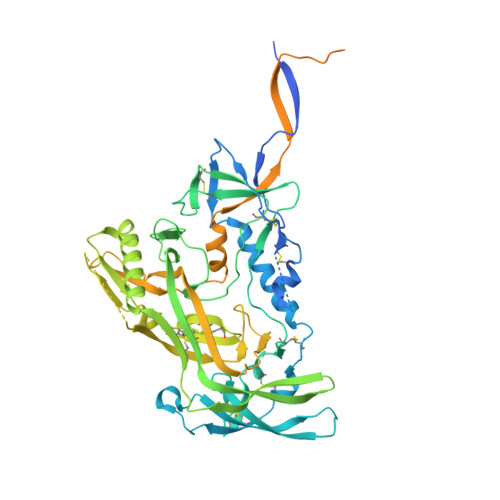
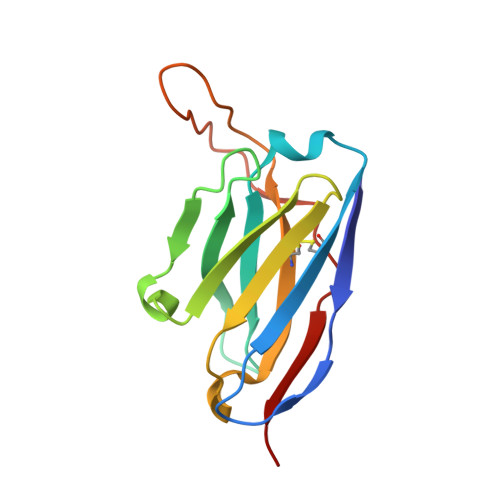
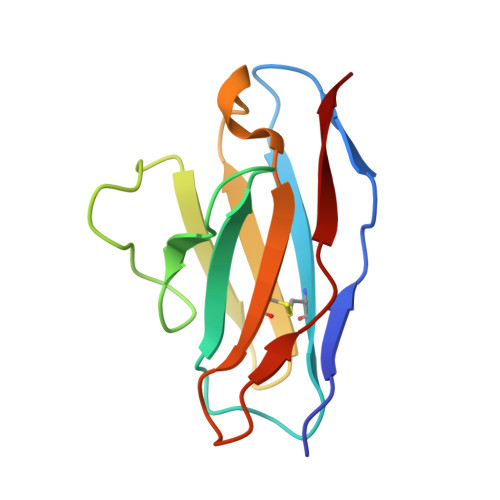
From structure to sequence: Antibody discovery using cryoEM.
Antanasijevic, A., Bowman, C.A., Kirchdoerfer, R.N., Cottrell, C.A., Ozorowski, G., Upadhyay, A.A., Cirelli, K.M., Carnathan, D.G., Enemuo, C.A., Sewall, L.M., Nogal, B., Zhao, F., Groschel, B., Schief, W.R., Sok, D., Silvestri, G., Crotty, S., Bosinger, S.E., Ward, A.B.(2022) Sci Adv 8: eabk2039-eabk2039
- PubMed: 35044813
- DOI: https://doi.org/10.1126/sciadv.abk2039
- Primary Citation of Related Structures:
7MDT, 7MDU, 7MEP - PubMed Abstract:
One of the rate-limiting steps in analyzing immune responses to vaccines or infections is the isolation and characterization of monoclonal antibodies. Here, we present a hybrid structural and bioinformatic approach to directly assign the heavy and light chains, identify complementarity-determining regions, and discover sequences from cryoEM density maps of serum-derived polyclonal antibodies bound to an antigen. When combined with next-generation sequencing of immune repertoires, we were able to specifically identify clonal family members, synthesize the monoclonal antibodies, and confirm that they interact with the antigen in a manner equivalent to the corresponding polyclonal antibodies. This structure-based approach for identification of monoclonal antibodies from polyclonal sera opens new avenues for analysis of immune responses and iterative vaccine design.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: