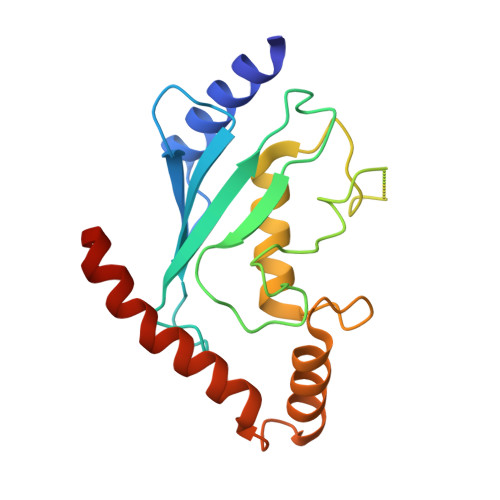
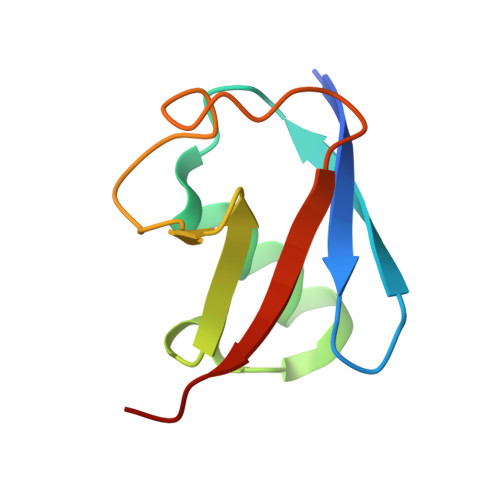
Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex.
St-Cyr, D., Ceccarelli, D.F., Orlicky, S., van der Sloot, A.M., Tang, X., Kelso, S., Moore, S., James, C., Posternak, G., Coulombe-Huntington, J., Bertomeu, T., Marinier, A., Sicheri, F., Tyers, M.(2021) Sci Adv 7: eabi5797-eabi5797
- PubMed: 34705497
- DOI: https://doi.org/10.1126/sciadv.abi5797
- Primary Citation of Related Structures:
7M2K - PubMed Abstract:
Pharmacological control of the ubiquitin-proteasome system (UPS) is of intense interest in drug discovery. Here, we report the development of chemical inhibitors of the ubiquitin-conjugating (E2) enzyme CDC34A (also known as UBE2R1), which donates activated ubiquitin to the cullin-RING ligase (CRL) family of ubiquitin ligase (E3) enzymes. A FRET-based interaction assay was used to screen for novel compounds that stabilize the noncovalent complex between CDC34A and ubiquitin, and thereby inhibit the CDC34A catalytic cycle. An isonipecotamide hit compound was elaborated into analogs with ~1000-fold increased potency in stabilizing the CDC34A-ubiquitin complex. These analogs specifically inhibited CDC34A-dependent ubiquitination in vitro and stabilized an E2~ubiquitin thioester reaction intermediate in cells. The x-ray crystal structure of a CDC34A-ubiquitin-inhibitor complex uncovered the basis for analog structure-activity relationships. The development of chemical stabilizers of the CDC34A-ubiquitin complex illustrates a general strategy for de novo discovery of molecular glue compounds that stabilize weak protein interactions.
- Institute for Research in Immunology and Cancer, University of Montreal, Montreal, Québec H3T 1J4, Canada.
Organizational Affiliation: