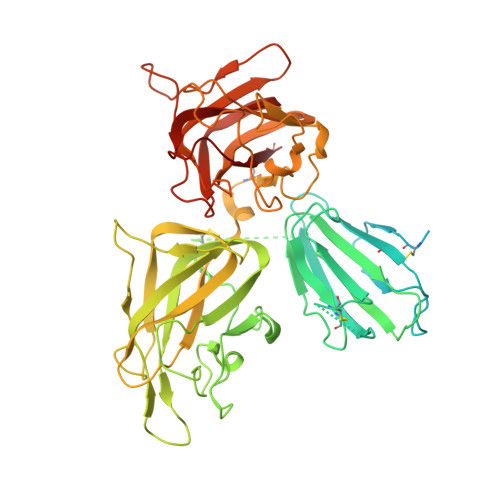
Structural basis for HCMV Pentamer recognition by neuropilin 2 and neutralizing antibodies.
Wrapp, D., Ye, X., Ku, Z., Su, H., Jones, H.G., Wang, N., Mishra, A.K., Freed, D.C., Li, F., Tang, A., Li, L., Jaijyan, D.K., Zhu, H., Wang, D., Fu, T.M., Zhang, N., An, Z., McLellan, J.S.(2022) Sci Adv 8: eabm2546-eabm2546
- PubMed: 35275718
- DOI: https://doi.org/10.1126/sciadv.abm2546
- Primary Citation of Related Structures:
7KBA, 7KBB, 7LYV, 7LYW, 7M1C, 7M22, 7M30 - PubMed Abstract:
Human cytomegalovirus (HCMV) encodes multiple surface glycoprotein complexes to infect a variety of cell types. The HCMV Pentamer, composed of gH, gL, UL128, UL130, and UL131A, enhances entry into epithelial, endothelial, and myeloid cells by interacting with the cell surface receptor neuropilin 2 (NRP2). Despite the critical nature of this interaction, the molecular determinants that govern NRP2 recognition remain unclear. Here, we describe the cryo-EM structure of NRP2 bound to Pentamer. The high-affinity interaction between these proteins is calcium dependent and differs from the canonical carboxyl-terminal arginine (CendR) binding that NRP2 typically uses. We also determine the structures of four neutralizing human antibodies bound to the HCMV Pentamer to define susceptible epitopes. Two of these antibodies compete with NRP2 binding, but the two most potent antibodies recognize a previously unidentified epitope that does not overlap the NRP2-binding site. Collectively, these findings provide a structural basis for HCMV tropism and antibody-mediated neutralization.
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.
Organizational Affiliation: