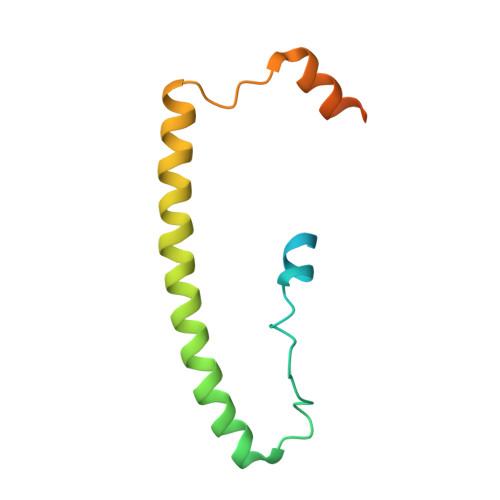
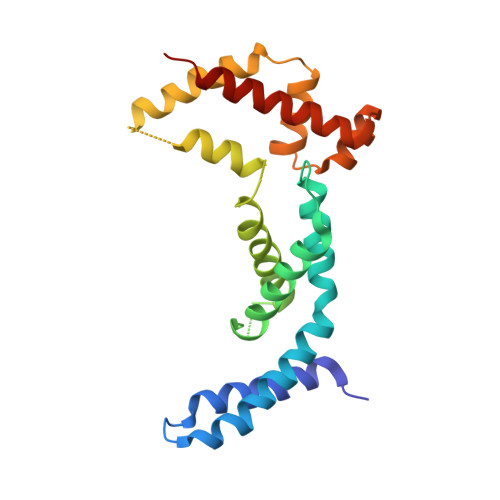
Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Schumacher, M.A., Gallagher, K.A., Holmes, N.A., Chandra, G., Henderson, M., Kysela, D.T., Brennan, R.G., Buttner, M.J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34290147
- DOI: https://doi.org/10.1073/pnas.2105447118
- Primary Citation of Related Structures:
7LQ2, 7LQ3, 7LQ4 - PubMed Abstract:
Filamentous actinobacteria of the genus Streptomyces have a complex lifecycle involving the differentiation of reproductive aerial hyphae into spores. We recently showed c-di-GMP controls this transition by arming a unique anti-σ, RsiG, to bind the sporulation-specific σ, WhiG. The Streptomyces venezuelae RsiG-(c-di-GMP) 2 -WhiG structure revealed that a monomeric RsiG binds c-di-GMP via two E(X) 3 S(X) 2 R(X) 3 Q(X) 3 D repeat motifs, one on each helix of an antiparallel coiled-coil. Here we show that RsiG homologs are found scattered throughout the Actinobacteria. Strikingly, RsiGs from unicellular bacteria descending from the most basal branch of the Actinobacteria are small proteins containing only one c-di-GMP binding motif, yet still bind their WhiG partners. Our structure of a Rubrobacter radiotolerans (RsiG) 2 -(c-di-GMP) 2 -WhiG complex revealed that these single-motif RsiGs are able to form an antiparallel coiled-coil through homodimerization, thereby allowing them to bind c-di-GMP similar to the monomeric twin-motif RsiGs. Further data show that in the unicellular actinobacterium R. radiotolerans , the (RsiG) 2 -(c-di-GMP) 2 -WhiG regulatory switch controls type IV pilus expression. Phylogenetic analysis indicates the single-motif RsiGs likely represent the ancestral state and an internal gene-duplication event gave rise to the twin-motif RsiGs inherited elsewhere in the Actinobacteria. Thus, these studies show how the anti-σ RsiG has evolved through an intragenic duplication event from a small protein carrying a single c-di-GMP binding motif, which functions as a homodimer, to a larger protein carrying two c-di-GMP binding motifs, which functions as a monomer. Consistent with this, our structures reveal potential selective advantages of the monomeric twin-motif anti-σ factors.
- Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710; maria.schumacher@duke.edu mark.buttner@jic.ac.uk.
Organizational Affiliation: