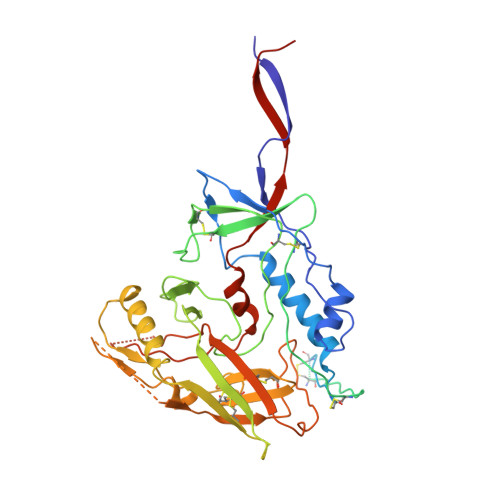
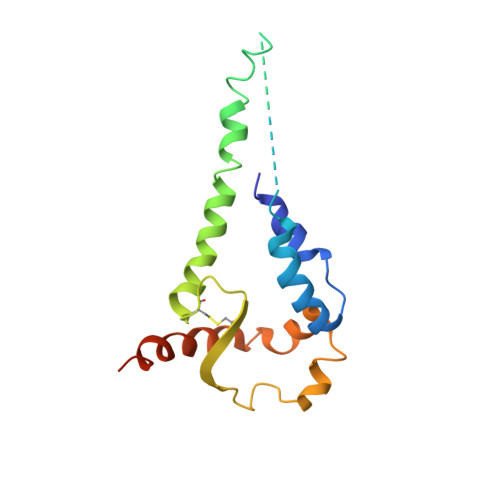
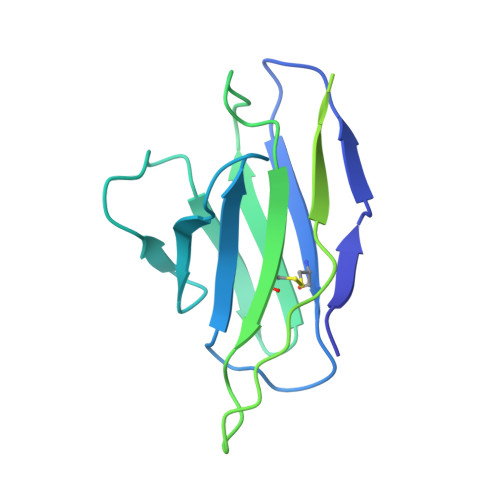
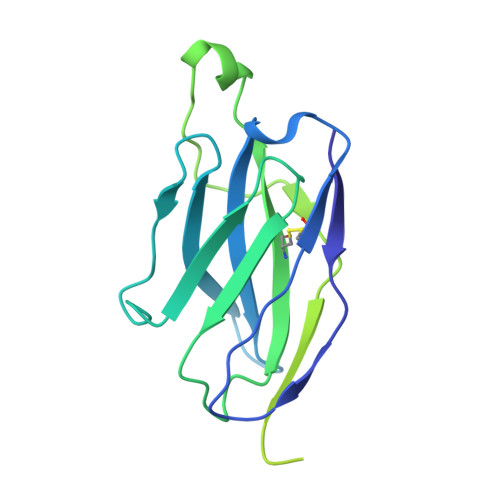
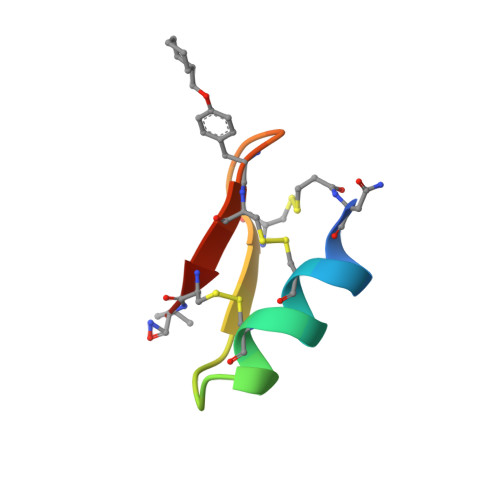
Cryo-EM structures of HIV-1 trimer bound to CD4-mimetics BNM-III-170 and M48U1 adopt a CD4-bound open conformation.
Jette, C.A., Barnes, C.O., Kirk, S.M., Melillo, B., Smith III, A.B., Bjorkman, P.J.(2021) Nat Commun 12: 1950-1950
- PubMed: 33782388
- DOI: https://doi.org/10.1038/s41467-021-21816-x
- Primary Citation of Related Structures:
7LO6, 7LOK - PubMed Abstract:
Human immunodeficiency virus-1 (HIV-1), the causative agent of AIDS, impacts millions of people. Entry into target cells is mediated by the HIV-1 envelope (Env) glycoprotein interacting with host receptor CD4, which triggers conformational changes allowing binding to a coreceptor and subsequent membrane fusion. Small molecule or peptide CD4-mimetic drugs mimic CD4's Phe43 interaction with Env by inserting into the conserved Phe43 pocket on Env subunit gp120. Here, we present single-particle cryo-EM structures of CD4-mimetics BNM-III-170 and M48U1 bound to a BG505 native-like Env trimer plus the CD4-induced antibody 17b at 3.7 Å and 3.9 Å resolution, respectively. CD4-mimetic-bound BG505 exhibits canonical CD4-induced conformational changes including trimer opening, formation of the 4-stranded gp120 bridging sheet, displacement of the V1V2 loop, and formation of a compact and elongated gp41 HR1C helical bundle. We conclude that CD4-induced structural changes on both gp120 and gp41 Env subunits are induced by binding to the gp120 Phe43 pocket.
- Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA, USA.
Organizational Affiliation: