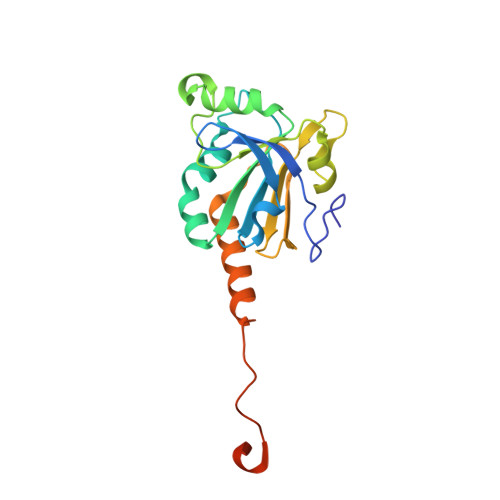
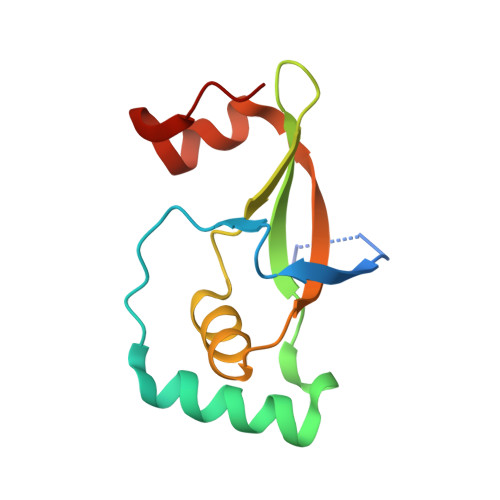
Specificity of Human Sulfiredoxin for Reductant and Peroxiredoxin Oligomeric State.
Forshaw, T.E., Reisz, J.A., Nelson, K.J., Gumpena, R., Lawson, J.R., Jonsson, T.J., Wu, H., Clodfelter, J.E., Johnson, L.C., Furdui, C.M., Lowther, W.T.(2021) Antioxidants (Basel) 10
- PubMed: 34208049
- DOI: https://doi.org/10.3390/antiox10060946
- Primary Citation of Related Structures:
7LJ1 - PubMed Abstract:
Human peroxiredoxins (Prx) are a family of antioxidant enzymes involved in a myriad of cellular functions and diseases. During the reaction with peroxides (e.g., H 2 O 2 ), the typical 2-Cys Prxs change oligomeric structure between higher order (do)decamers and disulfide-linked dimers, with the hyperoxidized inactive state (-SO 2 H) favoring the multimeric structure of the reduced enzyme. Here, we present a study on the structural requirements for the repair of hyperoxidized 2-Cys Prxs by human sulfiredoxin (Srx) and the relative efficacy of physiological reductants hydrogen sulfide (H 2 S) and glutathione (GSH) in this reaction. The crystal structure of the toroidal Prx1-Srx complex shows an extended active site interface. The loss of this interface within engineered Prx2 and Prx3 dimers yielded variants more resistant to hyperoxidation and repair by Srx. Finally, we reveal for the first time Prx isoform-dependent use of and potential cooperation between GSH and H 2 S in supporting Srx activity.
- Department of Internal Medicine, Section on Molecular Medicine, Wake Forest School of Medicine, Medical Center Blvd., Winston-Salem, NC 27157, USA.
Organizational Affiliation: