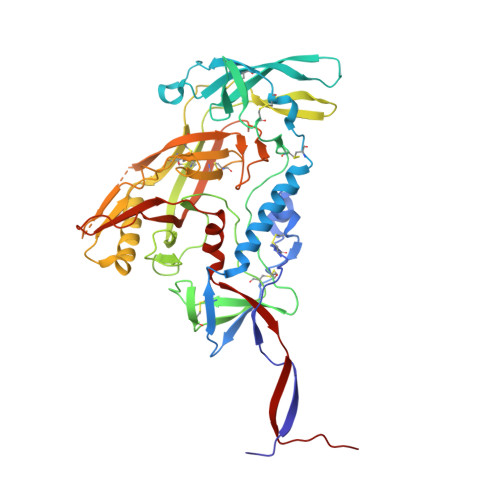
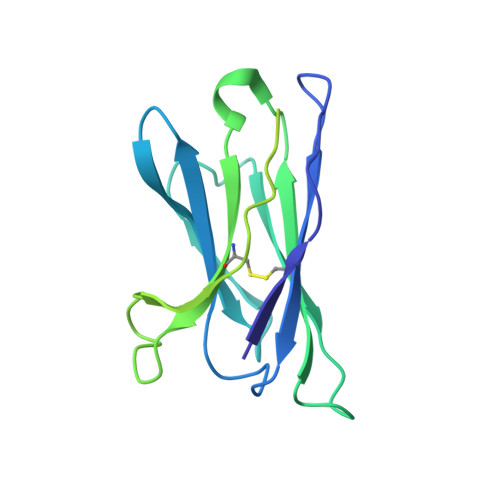
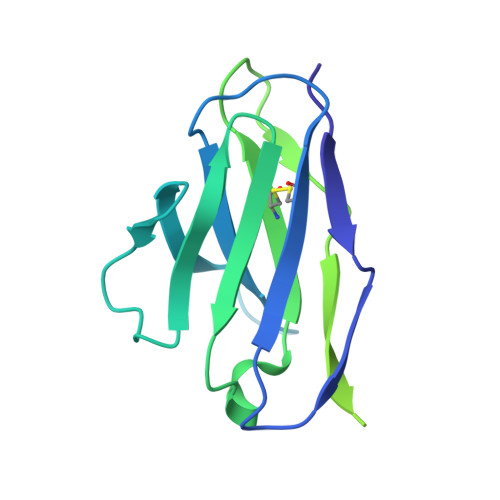
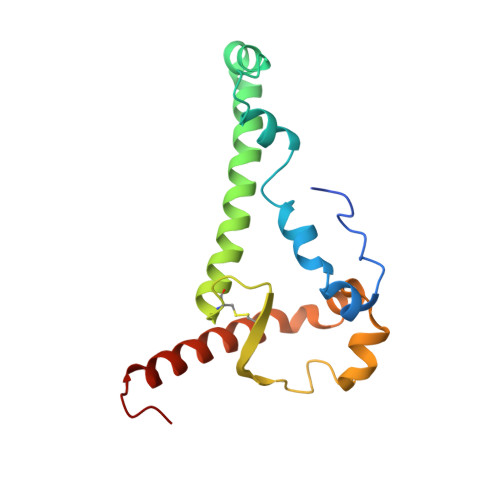
Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cottrell, C.A., Manne, K., Kong, R., Wang, S., Zhou, T., Chuang, G.Y., Edwards, R.J., Henderson, R., Janowska, K., Kopp, M., Lin, B.C., Louder, M.K., Olia, A.S., Rawi, R., Shen, C.H., Taft, J.D., Torres, J.L., Wu, N.R., Zhang, B., Doria-Rose, N.A., Cohen, M.S., Haynes, B.F., Shapiro, L., Ward, A.B., Acharya, P., Mascola, J.R., Kwong, P.D.(2021) Cell Rep 37: 109922-109922
- PubMed: 34731616
- DOI: https://doi.org/10.1016/j.celrep.2021.109922
- Primary Citation of Related Structures:
7LG6, 7LL1, 7LL2, 7LLK - PubMed Abstract:
Recognition of N-linked glycan at residue N276 (glycan276) at the periphery of the CD4-binding site (CD4bs) on the HIV-envelope trimer is a formidable challenge for many CD4bs-directed antibodies. To understand how this glycan can be recognized, here we isolate two lineages of glycan276-dependent CD4bs antibodies. Antibody CH540-VRC40.01 (named for donor-lineage.clone) neutralizes 81% of a panel of 208 diverse strains, while antibody CH314-VRC33.01 neutralizes 45%. Cryo-electron microscopy (cryo-EM) structures of these two antibodies and 179NC75, a previously identified glycan276-dependent CD4bs antibody, in complex with HIV-envelope trimer reveal substantially different modes of glycan276 recognition. Despite these differences, binding of glycan276-dependent antibodies maintains a glycan276 conformation similar to that observed in the absence of glycan276-binding antibodies. By contrast, glycan276-independent CD4bs antibodies, such as VRC01, displace glycan276 upon binding. These results provide a foundation for understanding antibody recognition of glycan276 and suggest its presence may be crucial for priming immunogens seeking to initiate broad CD4bs recognition.
- IAVI Neutralizing Antibody Center, Consortium for HIV/AIDS Vaccine Development (CHAVD), Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: