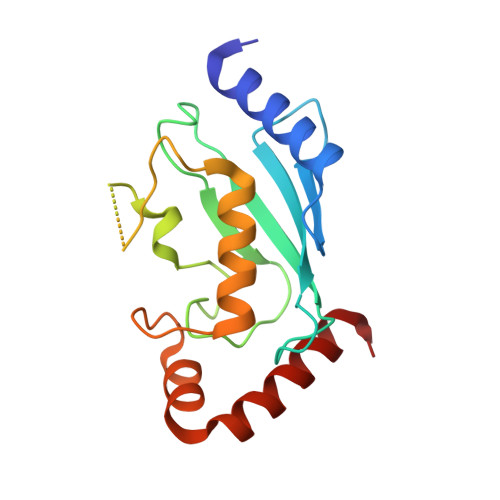
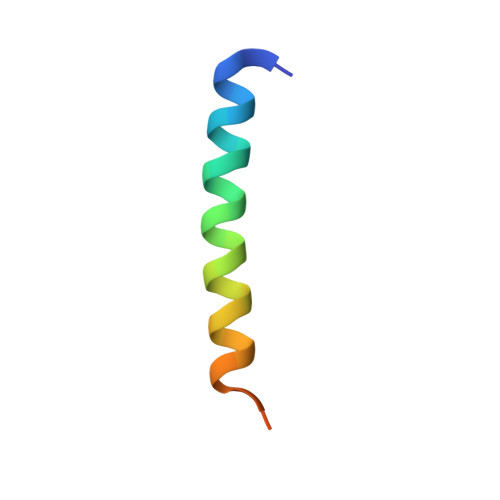
A structurally conserved site in AUP1 binds the E2 enzyme UBE2G2 and is essential for ER-associated degradation.
Smith, C.E., Tsai, Y.C., Liang, Y.H., Khago, D., Mariano, J., Li, J., Tarasov, S.G., Gergel, E., Tsai, B., Villaneuva, M., Clapp, M.E., Magidson, V., Chari, R., Byrd, R.A., Ji, X., Weissman, A.M.(2021) PLoS Biol 19: e3001474-e3001474
- PubMed: 34879065
- DOI: https://doi.org/10.1371/journal.pbio.3001474
- Primary Citation of Related Structures:
7LEW - PubMed Abstract:
Endoplasmic reticulum-associated degradation (ERAD) is a protein quality control pathway of fundamental importance to cellular homeostasis. Although multiple ERAD pathways exist for targeting topologically distinct substrates, all pathways require substrate ubiquitination. Here, we characterize a key role for the UBE2G2 Binding Region (G2BR) of the ERAD accessory protein ancient ubiquitous protein 1 (AUP1) in ERAD pathways. This 27-amino acid (aa) region of AUP1 binds with high specificity and low nanomolar affinity to the backside of the ERAD ubiquitin-conjugating enzyme (E2) UBE2G2. The structure of the AUP1 G2BR (G2BRAUP1) in complex with UBE2G2 reveals an interface that includes a network of salt bridges, hydrogen bonds, and hydrophobic interactions essential for AUP1 function in cells. The G2BRAUP1 shares significant structural conservation with the G2BR found in the E3 ubiquitin ligase gp78 and in vitro can similarly allosterically activate ubiquitination in conjunction with ERAD E3s. In cells, AUP1 is uniquely required to maintain normal levels of UBE2G2; this is due to G2BRAUP1 binding to the E2 and preventing its rapid degradation. In addition, the G2BRAUP1 is required for both ER membrane recruitment of UBE2G2 and for its activation at the ER membrane. Thus, by binding to the backside of a critical ERAD E2, G2BRAUP1 plays multiple critical roles in ERAD.
- Laboratory of Protein Dynamics and Signaling, Center for Cancer Research, NCI, National Institutes of Health, Frederick, Maryland, United States of America.
Organizational Affiliation: