Cryo-EM structure of the mature and infective Mayaro virus at 4.4 angstrom resolution reveals features of arthritogenic alphaviruses.
Ribeiro-Filho, H.V., Coimbra, L.D., Cassago, A., Rocha, R.P.F., Guerra, J.V.D.S., de Felicio, R., Carnieli, C.M., Leme, L., Padilha, A.C., Paes Leme, A.F., Trivella, D.B.B., Portugal, R.V., Lopes-de-Oliveira, P.S., Marques, R.E.(2021) Nat Commun 12: 3038-3038
- PubMed: 34031424
- DOI: https://doi.org/10.1038/s41467-021-23400-9
- Primary Citation of Related Structures:
7KO8 - PubMed Abstract:
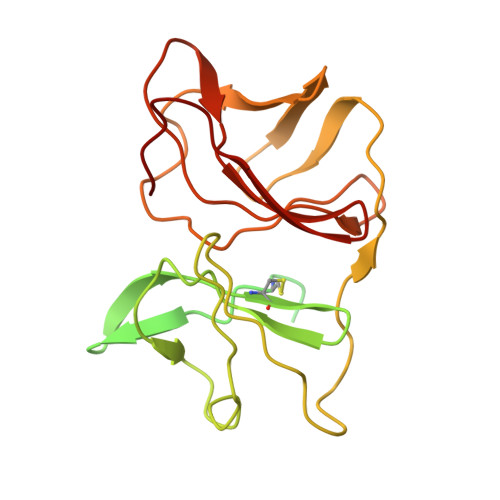
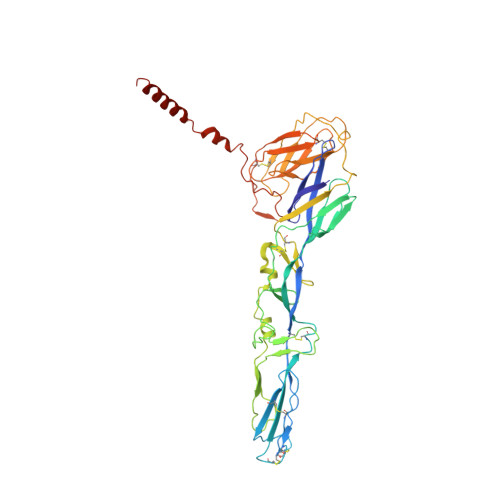
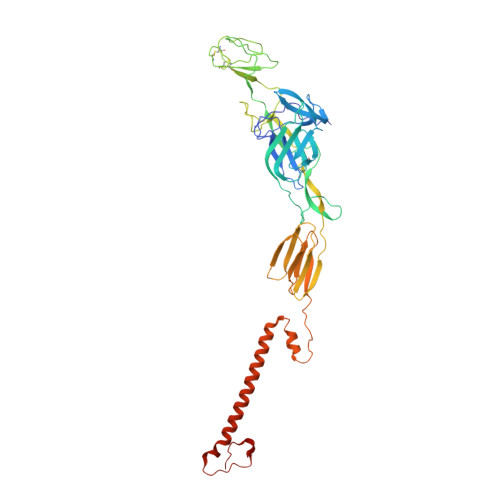
Mayaro virus (MAYV) is an emerging arbovirus of the Americas that may cause a debilitating arthritogenic disease. The biology of MAYV is not fully understood and largely inferred from related arthritogenic alphaviruses. Here, we present the structure of MAYV at 4.4 Å resolution, obtained from a preparation of mature, infective virions. MAYV presents typical alphavirus features and organization. Interactions between viral proteins that lead to particle formation are described together with a hydrophobic pocket formed between E1 and E2 spike proteins and conformational epitopes specific of MAYV. We also describe MAYV glycosylation residues in E1 and E2 that may affect MXRA8 host receptor binding, and a molecular "handshake" between MAYV spikes formed by N262 glycosylation in adjacent E2 proteins. The structure of MAYV is suggestive of structural and functional complexity among alphaviruses, which may be targeted for specificity or antiviral activity.
- Brazilian Biosciences National Laboratory, Brazilian Center for Research in Energy and Materials (CNPEM), Campinas, SP, Brazil.
Organizational Affiliation: