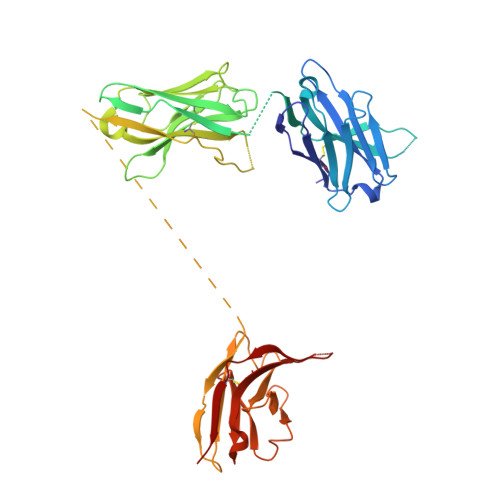
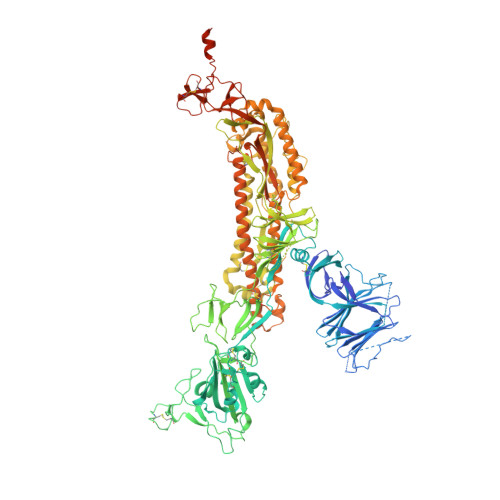
Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2.
Bracken, C.J., Lim, S.A., Solomon, P., Rettko, N.J., Nguyen, D.P., Zha, B.S., Schaefer, K., Byrnes, J.R., Zhou, J., Lui, I., Liu, J., Pance, K., Zhou, X.X., Leung, K.K., Wells, J.A.(2021) Nat Chem Biol 17: 113-121
- PubMed: 33082574
- DOI: https://doi.org/10.1038/s41589-020-00679-1
- Primary Citation of Related Structures:
7JWB - PubMed Abstract:
Neutralizing agents against SARS-CoV-2 are urgently needed for the treatment and prophylaxis of COVID-19. Here, we present a strategy to rapidly identify and assemble synthetic human variable heavy (VH) domains toward neutralizing epitopes. We constructed a VH-phage library and targeted the angiotensin-converting enzyme 2 (ACE2) binding interface of the SARS-CoV-2 Spike receptor-binding domain (Spike-RBD). Using a masked selection approach, we identified VH binders to two non-overlapping epitopes and further assembled these into multivalent and bi-paratopic formats. These VH constructs showed increased affinity to Spike (up to 600-fold) and neutralization potency (up to 1,400-fold) on pseudotyped SARS-CoV-2 virus when compared to standalone VH domains. The most potent binder, a trivalent VH, neutralized authentic SARS-CoV-2 with a half-maximal inhibitory concentration (IC 50 ) of 4.0 nM (180 ng ml -1 ). A cryo-EM structure of the trivalent VH bound to Spike shows each VH domain engaging an RBD at the ACE2 binding site, confirming our original design strategy.
- Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, CA, USA.
Organizational Affiliation: