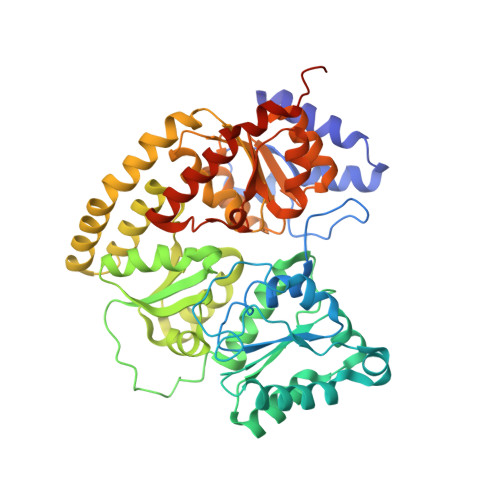
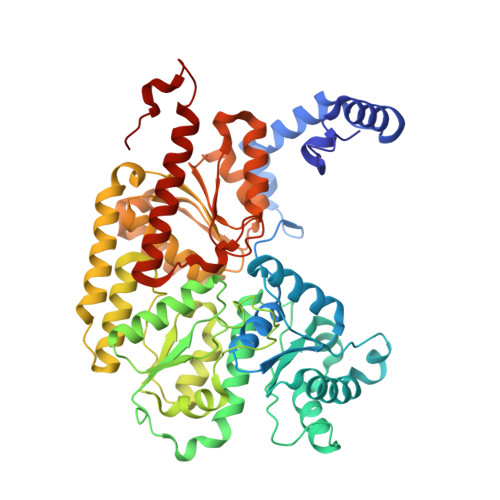
Structural Characterization of Two CO Molecules Bound to the Nitrogenase Active Site.
Buscagan, T.M., Perez, K.A., Maggiolo, A.O., Rees, D.C., Spatzal, T.(2021) Angew Chem Int Ed Engl 60: 5704-5707
- PubMed: 33320413
- DOI: https://doi.org/10.1002/anie.202015751
- Primary Citation of Related Structures:
7JRF - PubMed Abstract:
As an approach towards unraveling the nitrogenase mechanism, we have studied the binding of CO to the active-site FeMo-cofactor. CO is not only an inhibitor of nitrogenase, but it is also a substrate, undergoing reduction to hydrocarbons (Fischer-Tropsch-type chemistry). The C-C bond forming capabilities of nitrogenase suggest that multiple CO or CO-derived ligands bind to the active site. Herein, we report a crystal structure with two CO ligands coordinated to the FeMo-cofactor of the molybdenum nitrogenase at 1.33 Å resolution. In addition to the previously observed bridging CO ligand between Fe2 and Fe6 of the FeMo-cofactor, a new ligand binding mode is revealed through a second CO ligand coordinated terminally to Fe6. While the relevance of this state to nitrogenase-catalyzed reactions remains to be established, it highlights the privileged roles for Fe2 and Fe6 in ligand binding, with multiple coordination modes available depending on the ligand and reaction conditions.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA, 91125, USA.
Organizational Affiliation: