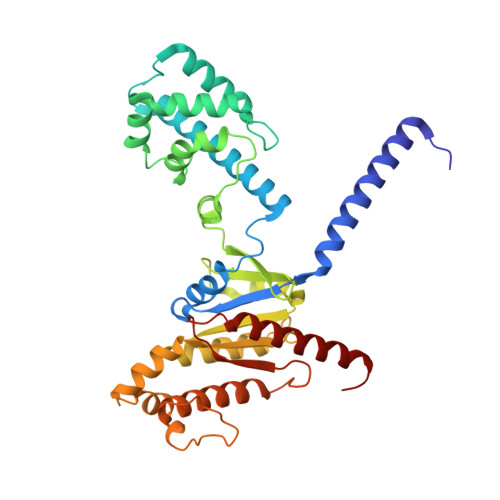
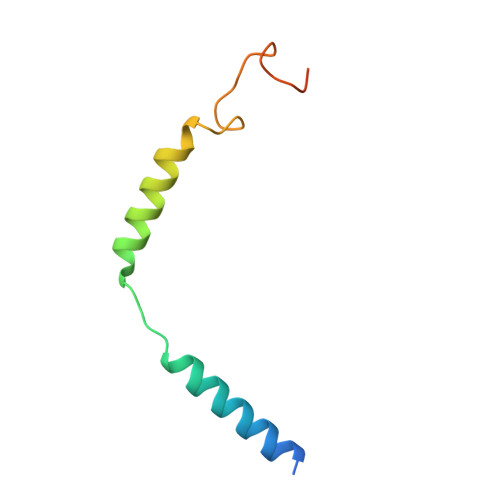
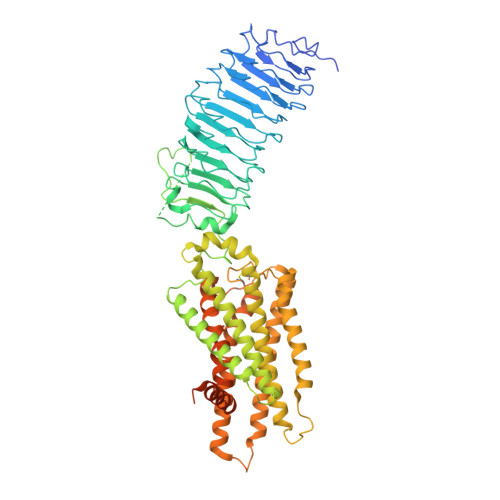
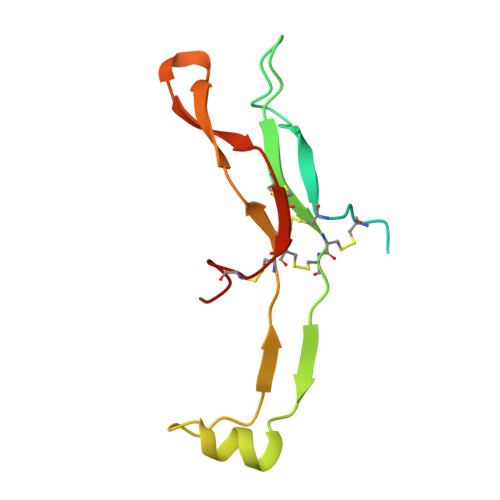
Structures of full-length glycoprotein hormone receptor signalling complexes.
Duan, J., Xu, P., Cheng, X., Mao, C., Croll, T., He, X., Shi, J., Luan, X., Yin, W., You, E., Liu, Q., Zhang, S., Jiang, H., Zhang, Y., Jiang, Y., Xu, H.E.(2021) Nature 598: 688-692
- PubMed: 34552239
- DOI: https://doi.org/10.1038/s41586-021-03924-2
- Primary Citation of Related Structures:
7FIG, 7FIH, 7FII, 7FIJ - PubMed Abstract:
Luteinizing hormone and chorionic gonadotropin are glycoprotein hormones that are related to follicle-stimulating hormone and thyroid-stimulating hormone 1,2 . Luteinizing hormone and chorionic gonadotropin are essential to human reproduction and are important therapeutic drugs 3-6 . They activate the same G-protein-coupled receptor, luteinizing hormone-choriogonadotropin receptor (LHCGR), by binding to the large extracellular domain 3 . Here we report four cryo-electron microscopy structures of LHCGR: two structures of the wild-type receptor in the inactive and active states; and two structures of the constitutively active mutated receptor. The active structures are bound to chorionic gonadotropin and the stimulatory G protein (G s ), and one of the structures also contains Org43553, an allosteric agonist 7 . The structures reveal a distinct 'push-and-pull' mechanism of receptor activation, in which the extracellular domain is pushed by the bound hormone and pulled by the extended hinge loop next to the transmembrane domain. A highly conserved 10-residue fragment (P10) from the hinge C-terminal loop at the interface between the extracellular domain and the transmembrane domain functions as a tethered agonist to induce conformational changes in the transmembrane domain and G-protein coupling. Org43553 binds to a pocket of the transmembrane domain and interacts directly with P10, which further stabilizes the active conformation. Together, these structures provide a common model for understanding the signalling of glycoprotein hormone receptors and a basis for drug discovery for endocrine diseases.
- The CAS Key Laboratory of Receptor Research and State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: