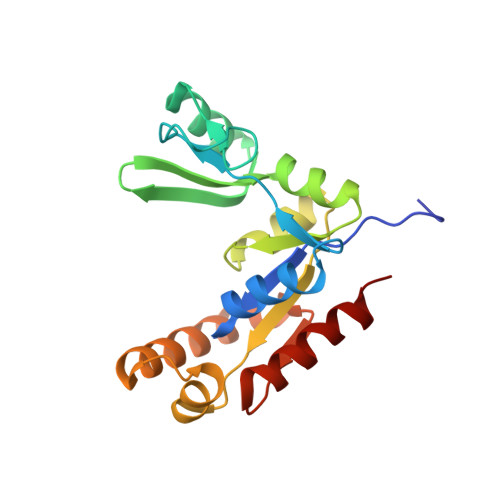
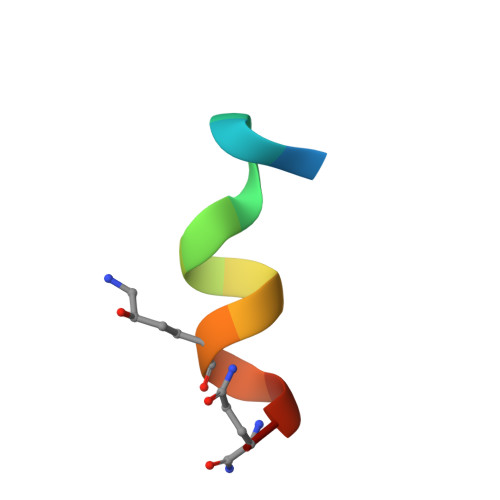
Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Unarta, I.C., Xu, J., Shang, Y., Cheung, C.H.P., Zhu, R., Chen, X., Cao, S., Cheung, P.P., Bierer, D., Zhang, M., Huang, X., Li, X.(2021) RSC Chem Biol 2: 1274-1284
- PubMed: 34458841
- DOI: https://doi.org/10.1039/d1cb00087j
- Primary Citation of Related Structures:
7F7G, 7F7I - PubMed Abstract:
Stapled peptides are promising protein-protein interaction (PPI) inhibitors that can increase the binding potency. Different from small-molecule inhibitors in which the binding mainly depends on energetic interactions with their protein targets, the binding of stapled peptides has long been suggested to be benefited from entropy. However, it remains challenging to reveal the molecular features that lead to this entropy gain, which could originate from the stabilization of the stapled peptide in solution or from the increased flexibility of the complex upon binding. This hinders the rational design of stapled peptides as PPI inhibitors. Using the guanylate kinase (GK) domain of the postsynaptic density protein 95 (PSD-95) as the target, we quantified the enthalpic and entropic contributions by combining isothermal titration calorimetry (ITC), X-ray crystallography, and free energy calculations based on all-atom molecular dynamics (MD) simulations. We successfully designed a stapled peptide inhibitor (staple 1) of the PSD-95 GK domain that led to a 25-fold increase in the binding affinity (from tens of μMs to 1.36 μM) with high cell permeability. We showed that entropy indeed greatly enhanced the binding affinity and the entropy gain was mainly due to the constrained-helix structure of the stapled peptide in solution (free state). Based on staple 1, we further designed two other stapled peptides (staple 2 and 3), which exerted even larger entropy gains compared to staple 1 because of their more flexible bound complexes (bound state). However, for staple 2 and 3, the overall binding affinities were not improved, as the loose binding in their bound states led to an enthalpic loss that largely compensated the excess entropy gain. Our work suggests that increasing the stability of the stapled peptide in free solution is an effective strategy for the rational design of stapled peptides as PPI inhibitors.
- Bioengineering Graduate Program, Department of Biological and Chemical Engineering, The Hong Kong University of Science and Technology, Clear Water Bay Kowloon Hong Kong xuhuihuang@ust.hk.
Organizational Affiliation: