Cryo-EM (SPA) structure of Nup155 N-terminus (19-863) at 5.2 Angstrom resolution
Niranjan, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| G protein/GFP fusion protein,Nuclear pore complex protein Nup155 | 1,114 | Recombinant vesicular stomatitis Indiana virus rVSV-G/GFP, Homo sapiens This entity is chimeric | Mutation(s): 0 Gene Names: G, GFP, NUP155, KIAA0791 | 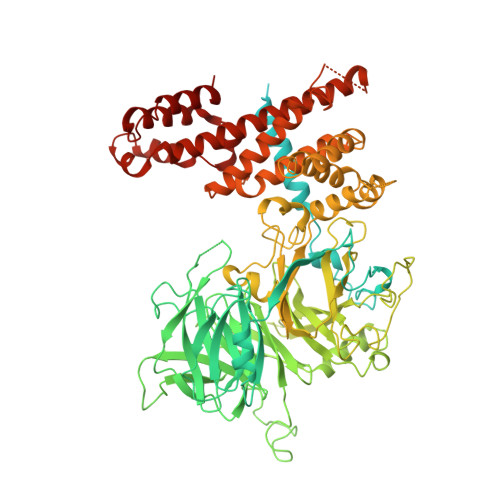 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for B7UCZ6 (Recombinant vesicular stomatitis Indiana virus rVSV-G/GFP) Explore B7UCZ6 Go to UniProtKB: B7UCZ6 | |||||
Find proteins for O75694 (Homo sapiens) Explore O75694 Go to UniProtKB: O75694 | |||||
PHAROS: O75694 GTEx: ENSG00000113569 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | B7UCZ6O75694 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Department of Biotechnology (DBT, India) | India | (DBT/PR/26398) |