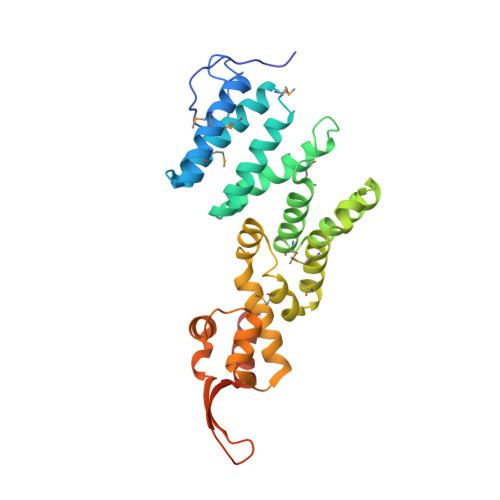
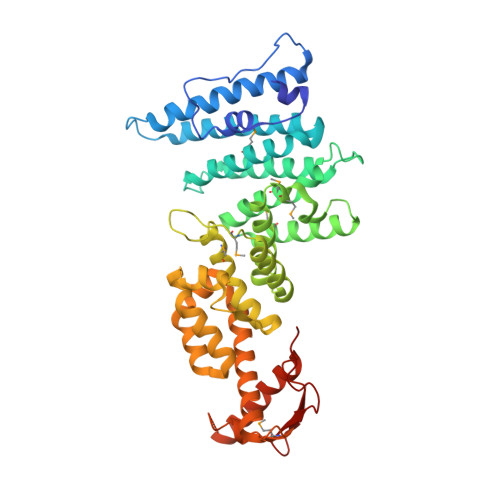
Structural assembly of the nucleic-acid-binding Thp3-Csn12-Sem1 complex functioning in mRNA splicing.
Kuang, Z., Ke, J., Hong, J., Zhu, Z., Niu, L.(2022) Nucleic Acids Res 50: 8882-8897
- PubMed: 35904806
- DOI: https://doi.org/10.1093/nar/gkac634
- Primary Citation of Related Structures:
7EWF, 7EWM - PubMed Abstract:
PCI domain proteins play important roles in post-transcriptional gene regulation. In the TREX-2 complex, PCI domain-containing Sac3 and Thp1 proteins and accessory Sem1 protein form a ternary complex required for mRNA nuclear export. In contrast, structurally related Thp3-Csn12-Sem1 complex mediates pre-mRNA splicing. In this study, we determined the structure of yeast Thp3186-470-Csn12-Sem1 ternary complex at 2.9 Å resolution. Both Thp3 and Csn12 structures have a typical PCI structural fold, characterized by a stack of α-helices capped by a C-terminal winged-helix (WH) domain. The overall structure of Thp3186-470-Csn12-Sem1 complex has an inverted V-shape with Thp3 and Csn12 forming the two sides. A fishhook-shaped Sem1 makes extensive contacts on Csn12 to stabilize its conformation. The overall structure of Thp3186-470-Csn12-Sem1 complex resembles the previously reported Sac3-Thp1-Sem1 complex, but also has significant structural differences. The C-terminal WH domains of Thp3 and Csn12 form a continuous surface to bind different forms of nucleic acids with micromolar affinity. Mutation of the basic residues in the WH domains of Thp3 and Csn12 affects nucleic acid binding in vitro and mRNA splicing in vivo. The Thp3-Csn12-Sem1 structure provides a foundation for further exploring the structural elements required for its specific recruitment to spliceosome for pre-mRNA splicing.
- Hefei National Laboratory for Physical Sciences at the Microscale, Division of Molecular and Cellular Biophysics, University of Science and Technology of China, Hefei, Anhui 230026, China.
Organizational Affiliation: