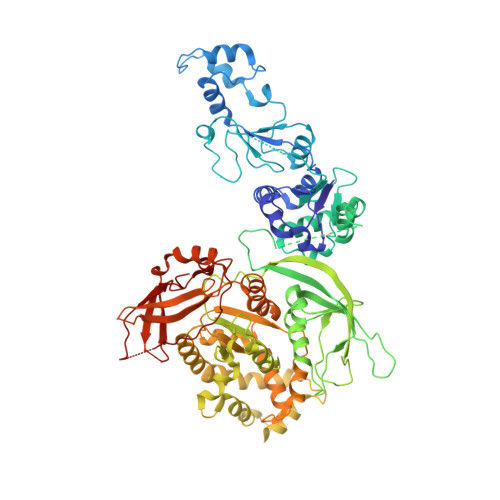
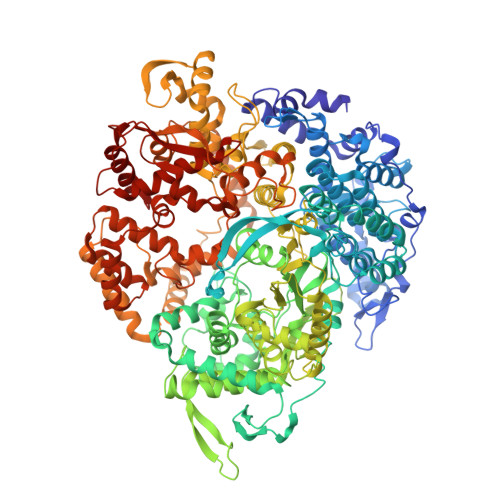
Asymmetric reconstruction of mammalian reovirus reveals interactions among RNA, transcriptional factor mu2 and capsid proteins.
Pan, M., Alvarez-Cabrera, A.L., Kang, J.S., Wang, L., Fan, C., Zhou, Z.H.(2021) Nat Commun 12: 4176-4176
- PubMed: 34234134
- DOI: https://doi.org/10.1038/s41467-021-24455-4
- Primary Citation of Related Structures:
7ELH, 7ELL - PubMed Abstract:
Mammalian reovirus (MRV) is the prototypical member of genus Orthoreovirus of family Reoviridae. However, lacking high-resolution structures of its RNA polymerase cofactor μ2 and infectious particle, limits understanding of molecular interactions among proteins and RNA, and their contributions to virion assembly and RNA transcription. Here, we report the 3.3 Å-resolution asymmetric reconstruction of transcribing MRV and in situ atomic models of its capsid proteins, the asymmetrically attached RNA-dependent RNA polymerase (RdRp) λ3, and RdRp-bound nucleoside triphosphatase μ2 with a unique RNA-binding domain. We reveal molecular interactions among virion proteins and genomic and messenger RNA. Polymerase complexes in three Spinoreovirinae subfamily members are organized with different pseudo-D 3d symmetries to engage their highly diversified genomes. The above interactions and those between symmetry-mismatched receptor-binding σ1 trimers and RNA-capping λ2 pentamers balance competing needs of capsid assembly, external protein removal, and allosteric triggering of endogenous RNA transcription, before, during and after infection, respectively.
- CAS Key Laboratory of Interfacial Physics and Technology, Shanghai Institute of Applied Physics, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: