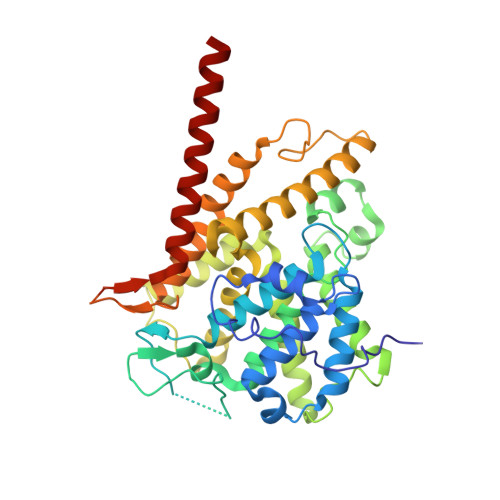
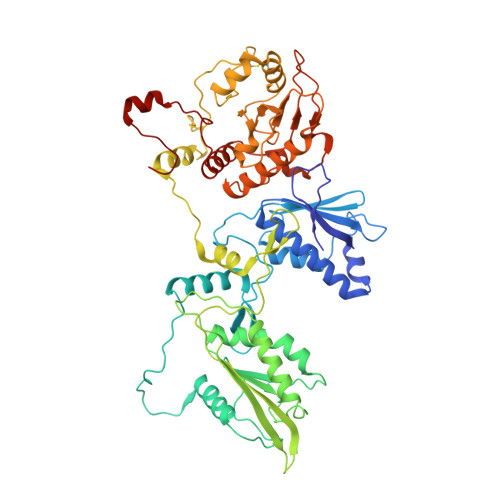
Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Chen, J., Liu, N., Huang, Y., Wang, Y., Sun, Y., Wu, Q., Li, D., Gao, S., Wang, H.W., Huang, N., Qi, X., Wang, X.(2021) Nat Commun 12: 6204-6204
- PubMed: 34707099
- DOI: https://doi.org/10.1038/s41467-021-26546-8
- Primary Citation of Related Structures:
7EG0, 7EG1, 7EG4 - PubMed Abstract:
Molecular glues are a class of small molecular drugs that mediate protein-protein interactions, that induce either the degradation or stabilization of target protein. A structurally diverse group of chemicals, including 17-β-estradiol (E2), anagrelide, nauclefine, and DNMDP, induces apoptosis by forming complexes with phosphodiesterase 3A (PDE3A) and Schlafen 12 protein (SLFN12). They do so by binding to the PDE3A enzymatic pocket that allows the compound-bound PDE3A to recruit and stabilize SLFN12, which in turn blocks protein translation, leading to apoptosis. In this work, we report the high-resolution cryo-electron microscopy structure of PDE3A-SLFN12 complexes isolated from cultured HeLa cells pre-treated with either anagrelide, or nauclefine, or DNMDP. The PDE3A-SLFN12 complexes exhibit a butterfly-like shape, forming a heterotetramer with these small molecules, which are packed in a shallow pocket in the catalytic domain of PDE3A. The resulting small molecule-modified interface binds to the short helix (E552-I558) of SLFN12 through hydrophobic interactions, thus "gluing" the two proteins together. Based on the complex structure, we designed and synthesized analogs of anagrelide, a known drug used for the treatment of thrombocytosis, to enhance their interactions with SLFN12, and achieved superior efficacy in inducing apoptosis in cultured cells as well as in tumor xenografts.
- National Institute of Biological Sciences, 7 Science Park Road, Zhongguancun Life Science Park, Beijing, 102206, China.
Organizational Affiliation: