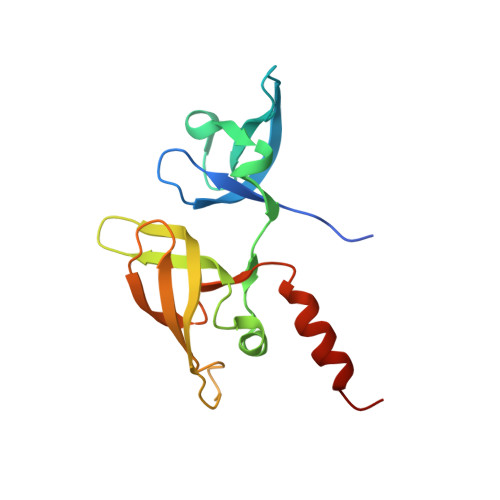
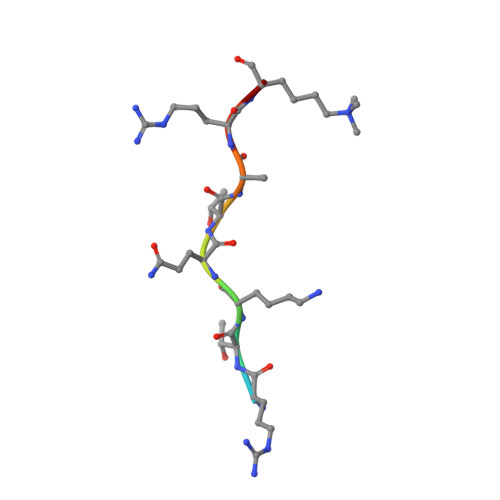
Recognition of H3K9me1 by maize RNA-directed DNA methylation factor SHH2.
Wang, Y., Zhou, X., Luo, J., Lv, S., Liu, R., Du, X., Jia, B., Yuan, F., Zhang, H., Du, J.(2021) J Integr Plant Biol 63: 1091-1096
- PubMed: 33913587
- DOI: https://doi.org/10.1111/jipb.13103
- Primary Citation of Related Structures:
7EEZ, 7EF0, 7EF1, 7EF2, 7EF3 - PubMed Abstract:
RNA-directed DNA methylation (RdDM) is a plant-specific de novo DNA methylation pathway, which has extensive cross-talk with histone modifications. Here, we report that the maize RdDM regulator SAWADEE HOMEODOMAIN HOMOLOG 2 (SHH2) is an H3K9me1 reader. Our structural studies reveal that H3K9me1 recognition is achieved by recognition of the methyl group via a classic aromatic cage and hydrogen-bonding and salt-bridge interactions with the free protons of the mono-methyllysine. The di- and tri-methylation states disrupt the polar interactions, decreasing the binding affinity. Our study reveals a mono-methyllysine recognition mechanism which potentially links RdDM to H3K9me1 in maize.
- National Key Laboratory of Plant Molecular Genetics and Shanghai Center for Plant Stress Biology, Center for Excellence in Molecular Plant Sciences, The Chinese Academy of Sciences, Shanghai, 201602, China.
Organizational Affiliation: