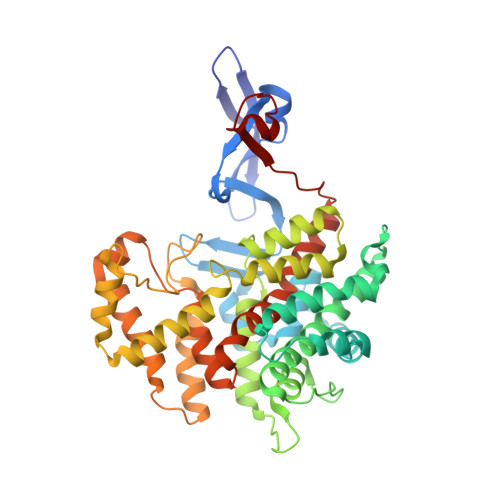
Structural basis of the cooperative activation of type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554.
Park, S.H., Lee, C.W., Bae, D.W., Do, H., Jeong, C.S., Hwang, J., Cha, S.S., Lee, J.H.(2021) Int J Biol Macromol 183: 213-221
- PubMed: 33910038
- DOI: https://doi.org/10.1016/j.ijbiomac.2021.04.141
- Primary Citation of Related Structures:
7E8N - PubMed Abstract:
Citrate synthase (CS) catalyzes the formation of citrate and coenzyme A from acetyl-CoA and oxaloacetate. CS exists in two forms: type I and type II. We determined the citrate-bound crystal structure of type II CS from the Hymenobacter sp. PAMC 26554 bacterium (HyCS; isolated from Antarctic lichen). Citrate molecules bound to a cleft between the large and small domains of HyCS. Structural comparison of HyCS with other type II CSs revealed that type II CSs have a highly conserved flexible hinge region (residues G264-P265 in HyCS), enabling correct positioning of active site residues. Notably, the catalytic His266 residue of HyCS interacted with Trp262 in the inactive (unliganded open) state of other type II CSs, whereas the His266 residue moved to the active site via a small-domain swing motion, interacting with the bound citrate in the closed conformation of HyCS. However, type I CSs lack this tryptophan residue and face-to-edge interactions. Thus, type II CSs might have a unique domain-motion control mechanism enabling a tight allosteric regulation. An activity assay using a W262A mutant showed a Hill coefficient of 2.4; thus, the interaction between Trp262 and His266 was closely related to the positive cooperative ligand binding of type II CS.
- Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Republic of Korea.
Organizational Affiliation: