Extraordinarily fast growth of high-quality glucose isomerase crystals simply by concentration in a precipitant-free solution with a cryoprotectant
Suzuki, Y., Ikemitsu, N., Maita, N.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Xylose isomerase | 386 | Streptomyces pseudogriseolus | Mutation(s): 1 EC: 5.3.1.5 | 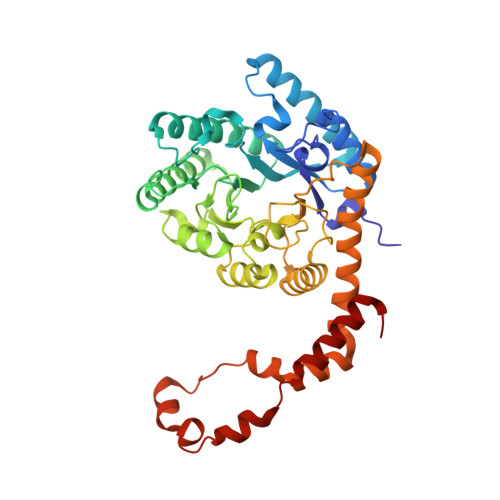 | |
UniProt | |||||
Find proteins for P24300 (Streptomyces rubiginosus) Explore P24300 Go to UniProtKB: P24300 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P24300 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | B [auth A], C [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| MN Query on MN | E [auth A], F [auth A] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| CA Query on CA | D [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.61 | α = 90 |
| b = 98.46 | β = 90 |
| c = 102.3 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Aimless | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 18K04960 |