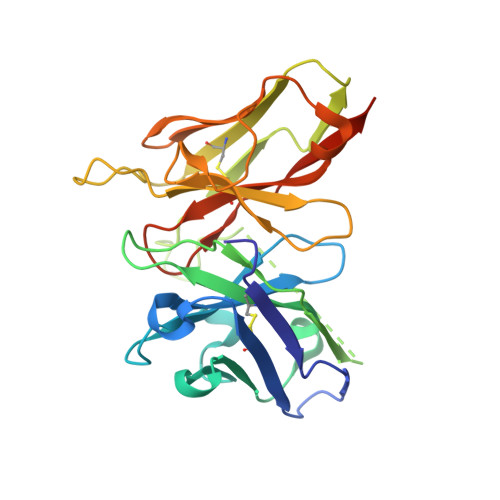
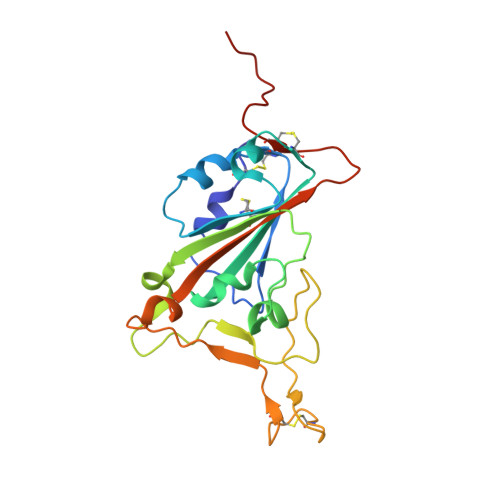
Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Fu, D., Zhang, G., Wang, Y., Zhang, Z., Hu, H., Shen, S., Wu, J., Li, B., Li, X., Fang, Y., Liu, J., Wang, Q., Zhou, Y., Wang, W., Li, Y., Lu, Z., Wang, X., Nie, C., Tian, Y., Chen, D., Wang, Y., Zhou, X., Wang, Q., Yu, F., Zhang, C., Deng, C., Zhou, L., Guan, G., Shao, N., Lou, Z., Deng, F., Zhang, H., Chen, X., Wang, M., Liu, L., Rao, Z., Guo, Y.(2021) PLoS Biol 19: e3001209-e3001209
- PubMed: 33961621
- DOI: https://doi.org/10.1371/journal.pbio.3001209
- Primary Citation of Related Structures:
7DEO, 7DET, 7DEU - PubMed Abstract:
The ongoing Coronavirus Disease 2019 (COVID-19) pandemic caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) threatens global public health and economy unprecedentedly, requiring accelerating development of prophylactic and therapeutic interventions. Molecular understanding of neutralizing antibodies (NAbs) would greatly help advance the development of monoclonal antibody (mAb) therapy, as well as the design of next generation recombinant vaccines. Here, we applied H2L2 transgenic mice encoding the human immunoglobulin variable regions, together with a state-of-the-art antibody discovery platform to immunize and isolate NAbs. From a large panel of isolated antibodies, 25 antibodies showed potent neutralizing activities at sub-nanomolar levels by engaging the spike receptor-binding domain (RBD). Importantly, one human NAb, termed PR1077, from the H2L2 platform and 2 humanized NAb, including PR953 and PR961, were further characterized and subjected for subsequent structural analysis. High-resolution X-ray crystallography structures unveiled novel epitopes on the receptor-binding motif (RBM) for PR1077 and PR953, which directly compete with human angiotensin-converting enzyme 2 (hACE2) for binding, and a novel non-blocking epitope on the neighboring site near RBM for PR961. Moreover, we further tested the antiviral efficiency of PR1077 in the Ad5-hACE2 transduction mouse model of COVID-19. A single injection provided potent protection against SARS-CoV-2 infection in either prophylactic or treatment groups. Taken together, these results shed light on the development of mAb-related therapeutic interventions for COVID-19.
- State Key Laboratory of Medicinal Chemical Biology and College of Pharmacy, Nankai University, Tianjin, China.
Organizational Affiliation: