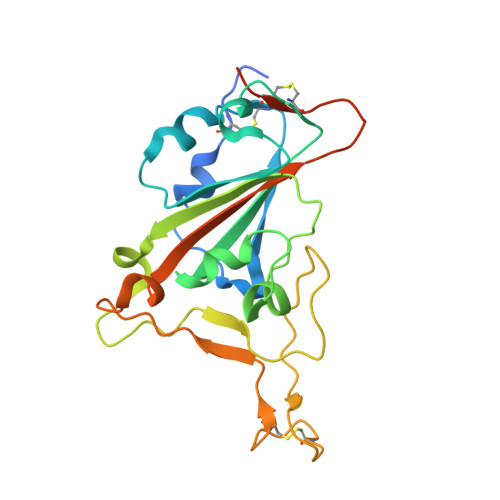
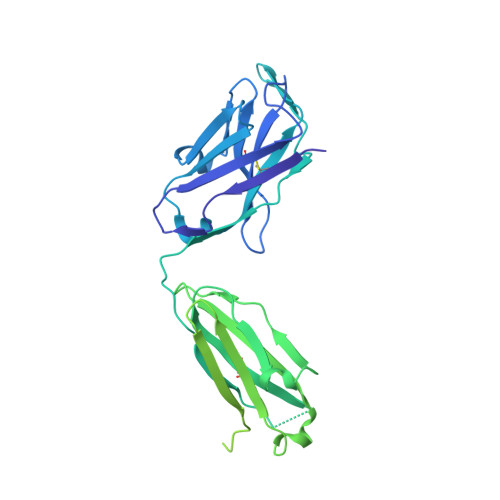
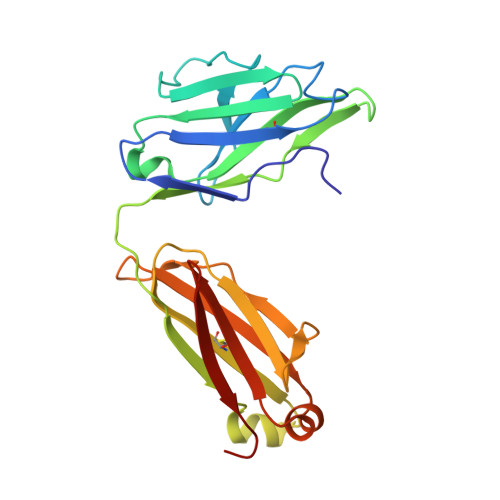
A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Kim, C., Ryu, D.K., Lee, J., Kim, Y.I., Seo, J.M., Kim, Y.G., Jeong, J.H., Kim, M., Kim, J.I., Kim, P., Bae, J.S., Shim, E.Y., Lee, M.S., Kim, M.S., Noh, H., Park, G.S., Park, J.S., Son, D., An, Y., Lee, J.N., Kwon, K.S., Lee, J.Y., Lee, H., Yang, J.S., Kim, K.C., Kim, S.S., Woo, H.M., Kim, J.W., Park, M.S., Yu, K.M., Kim, S.M., Kim, E.H., Park, S.J., Jeong, S.T., Yu, C.H., Song, Y., Gu, S.H., Oh, H., Koo, B.S., Hong, J.J., Ryu, C.M., Park, W.B., Oh, M.D., Choi, Y.K., Lee, S.Y.(2021) Nat Commun 12: 288-288
- PubMed: 33436577
- DOI: https://doi.org/10.1038/s41467-020-20602-5
- Primary Citation of Related Structures:
7CM4 - PubMed Abstract:
Vaccines and therapeutics are urgently needed for the pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here, we screen human monoclonal antibodies (mAb) targeting the receptor binding domain (RBD) of the viral spike protein via antibody library constructed from peripheral blood mononuclear cells of a convalescent patient. The CT-P59 mAb potently neutralizes SARS-CoV-2 isolates including the D614G variant without antibody-dependent enhancement effect. Complex crystal structure of CT-P59 Fab/RBD shows that CT-P59 blocks interaction regions of RBD for angiotensin converting enzyme 2 (ACE2) receptor with an orientation that is notably different from previously reported RBD-targeting mAbs. Furthermore, therapeutic effects of CT-P59 are evaluated in three animal models (ferret, hamster, and rhesus monkey), demonstrating a substantial reduction in viral titer along with alleviation of clinical symptoms. Therefore, CT-P59 may be a promising therapeutic candidate for COVID-19.
- Biotechnology Research Institute, Celltrion Inc, Incheon, 22014, Republic of Korea.
Organizational Affiliation: