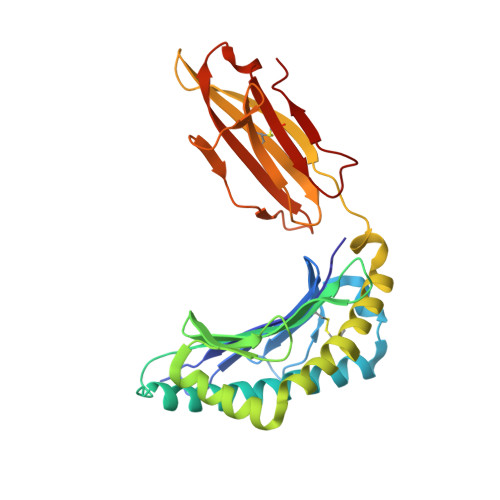
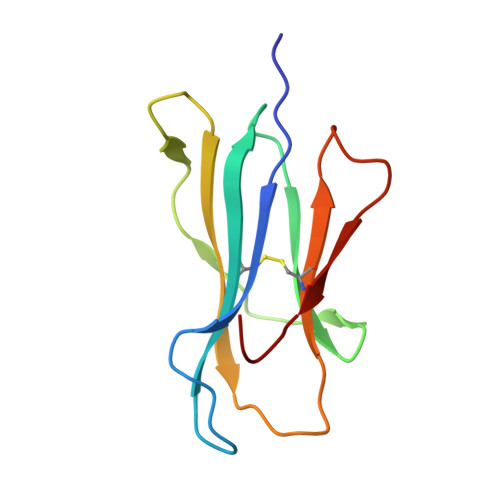
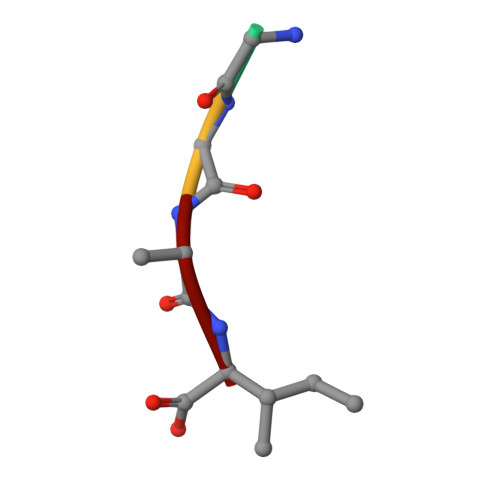
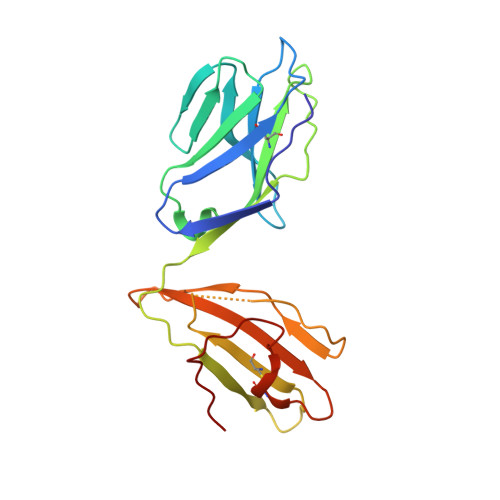
Crystal structure of the ternary complex of TCR, MHC class I and lipopeptides.
Morita, D., Iwashita, C., Mizutani, T., Mori, N., Mikami, B., Sugita, M.(2020) Int Immunol 32: 805-810
- PubMed: 32720986
- DOI: https://doi.org/10.1093/intimm/dxaa050
- Primary Citation of Related Structures:
7BYD - PubMed Abstract:
The covalent conjugation of a 14-carbon fatty acid (myristic acid) to the N-terminal Gly residue, termed N-myristoylation, occurs in some viral proteins to dictate their pathological function. This protein lipidation reaction, however, is monitored by host cytotoxic T lymphocytes that are capable of recognizing N-terminal lipopeptide fragments in the context of major histocompatibility complex (MHC) class I molecules. In a rhesus model of human AIDS, for example, the classical MHC class I allomorph, Mamu-B*05104, was shown to bind SIV Nef-derived 4-mer lipopeptides (myristic acid-Gly-Gly-Ala-Ile; C14nef4) and present them to the CD8+ T-cell line, SN45. These lipopeptides accommodated in MHC class I molecules expose much shorter peptide chains than conventional MHC class I-presented 8-10-mer peptides, and the molecular mechanisms by which αβ T-cell receptors (TCRs) recognize lipopeptides currently remain unclear. An X-ray crystallographic analysis of the SN45 TCR α and β heterodimer in a form that was co-crystallized with the C14nef4-bound Mamu-B*05104 complex indicated that the amide group of the N-myristoylated glycine residue offered a primary T-cell epitope by establishing a sole hydrogen bond between its nitrogen atom and the side chain of Glu at position 101 of CDR3β. Accordingly, the Glu to Ala mutation at this position resulted in the loss of lipopeptide recognition. On the other hand, TCRs were positioned remotely from the peptide portion of C14nef4, and strong interactions were not observed. Thus, these observations provide novel structural insights into lipopeptide recognition by TCRs, which contrast sharply with the general molecular principle of peptide recognition.
- Laboratory of Cell Regulation, Institute for Frontier Life and Medical Sciences, Kyoto University, Kawahara-cho, Shogoin, Sakyo-ku, Kyoto, Japan.
Organizational Affiliation: