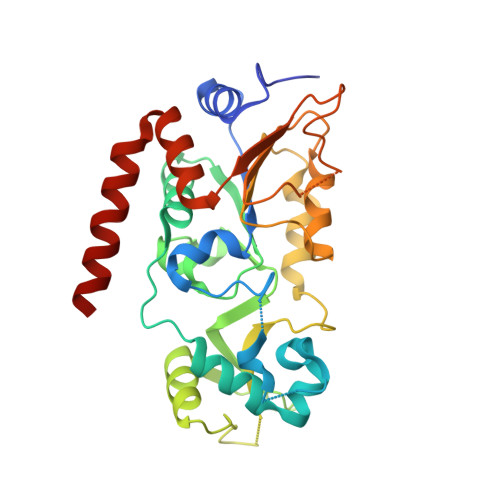
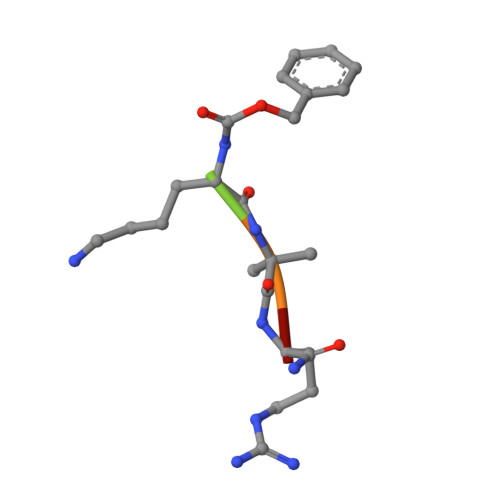
Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Nielsen, A.L., Rajabi, N., Kudo, N., Lundo, K., Moreno-Yruela, C., Baek, M., Fontenas, M., Lucidi, A., Madsen, A.S., Yoshida, M., Olsen, C.A.(2021) RSC Chem Biol 2: 612-626
- PubMed: 34458803
- DOI: https://doi.org/10.1039/d0cb00036a
- Primary Citation of Related Structures:
7BOS, 7BOT - PubMed Abstract:
Sirtuin 2 (SIRT2) is a protein deacylase enzyme that removes acetyl groups and longer chain acyl groups from post-translationally modified lysine residues. It affects diverse biological functions in the cell and has been considered a drug target in relation to both neurodegenerative diseases and cancer. Therefore, access to well-characterized and robust tool compounds is essential for the continued investigation of the complex functions of this enzyme. Here, we report a collection of chemical probes that are potent, selective, stable in serum, water-soluble, and inhibit SIRT2-mediated deacetylation and demyristoylation in cells. Compared to the current landscape of SIRT2 inhibitors, this is a unique ensemble of features built into a single compound. We expect the developed chemotypes to find broad application in the interrogation of SIRT2 functions in both healthy and diseased cells, and to provide a foundation for the development of future therapeutics.
- Center for Biopharmaceuticals & Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen Universitetsparken 2 DK-2100 Copenhagen Denmark cao@sund.ku.dk.
Organizational Affiliation: