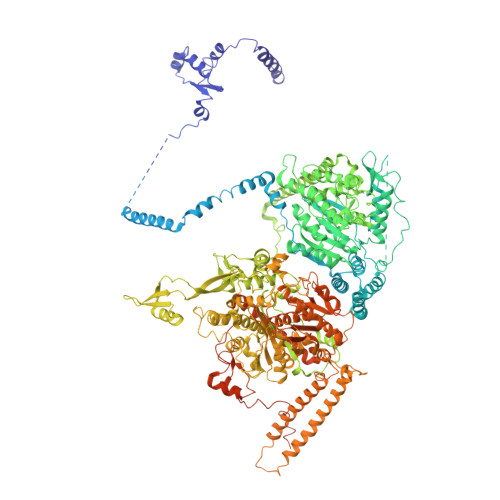
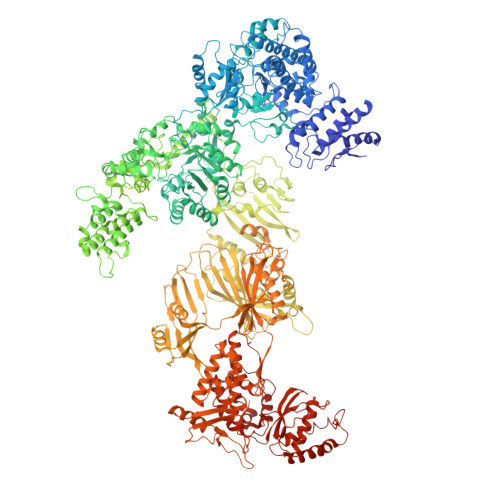
Structural insight into Pichia pastoris fatty acid synthase.
Snowden, J.S., Alzahrani, J., Sherry, L., Stacey, M., Rowlands, D.J., Ranson, N.A., Stonehouse, N.J.(2021) Sci Rep 11: 9773-9773
- PubMed: 33963233
- DOI: https://doi.org/10.1038/s41598-021-89196-2
- Primary Citation of Related Structures:
7BC4, 7BC5 - PubMed Abstract:
Type I fatty acid synthases (FASs) are critical metabolic enzymes which are common targets for bioengineering in the production of biofuels and other products. Serendipitously, we identified FAS as a contaminant in a cryoEM dataset of virus-like particles (VLPs) purified from P. pastoris, an important model organism and common expression system used in protein production. From these data, we determined the structure of P. pastoris FAS to 3.1 Å resolution. While the overall organisation of the complex was typical of type I FASs, we identified several differences in both structural and enzymatic domains through comparison with the prototypical yeast FAS from S. cerevisiae. Using focussed classification, we were also able to resolve and model the mobile acyl-carrier protein (ACP) domain, which is key for function. Ultimately, the structure reported here will be a useful resource for further efforts to engineer yeast FAS for synthesis of alternate products.
- Astbury Centre for Structural Molecular Biology, School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, LS2 9JT, UK.
Organizational Affiliation: