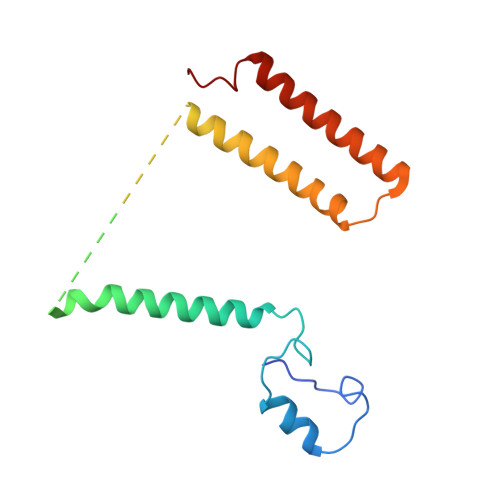
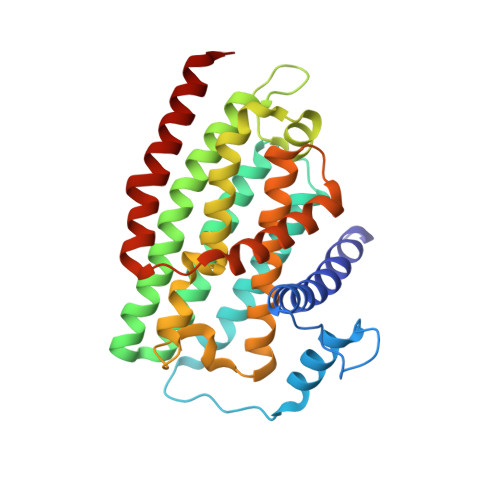
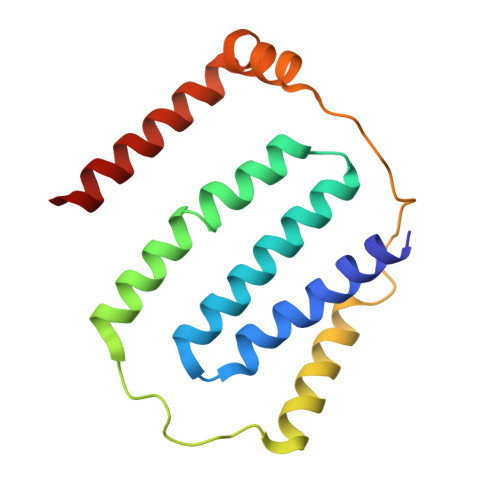
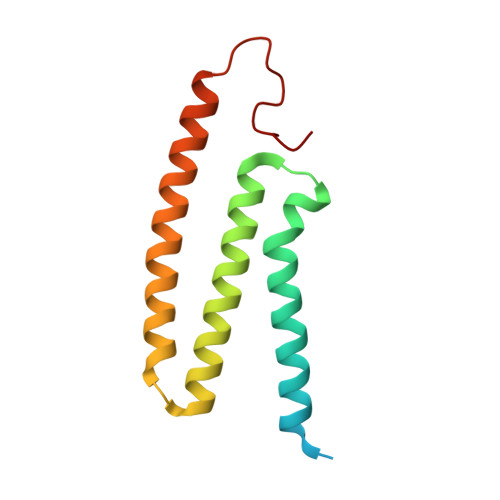
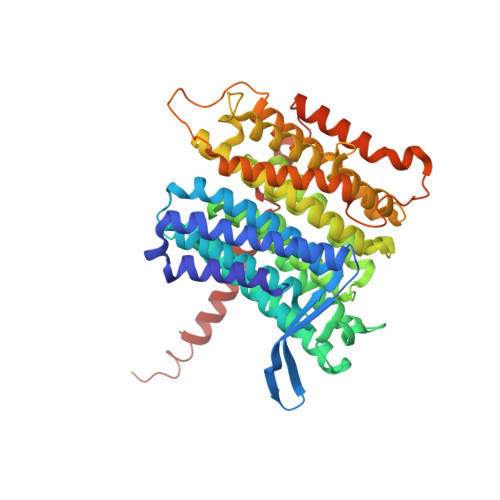
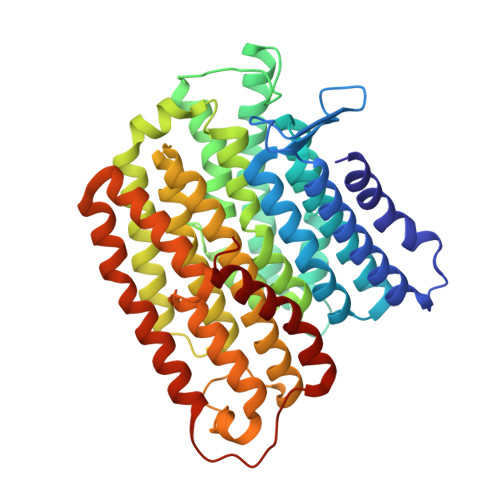
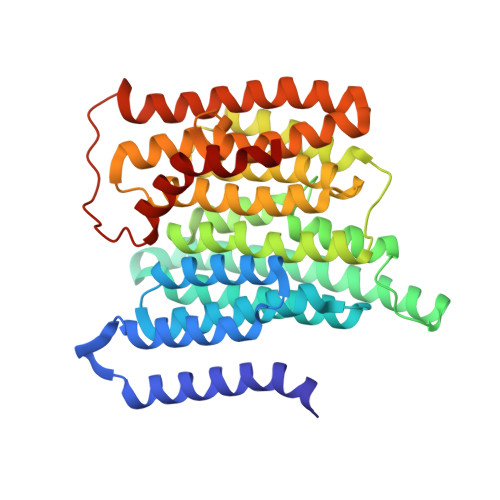
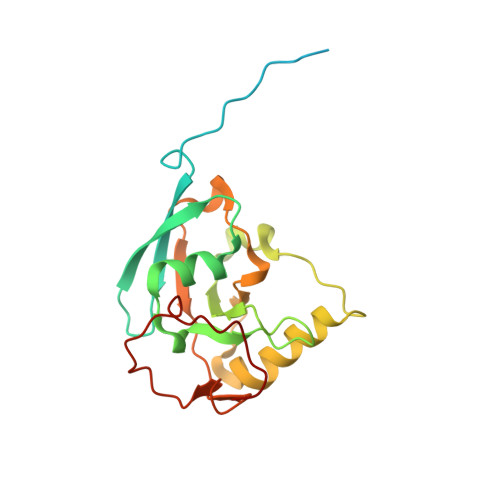
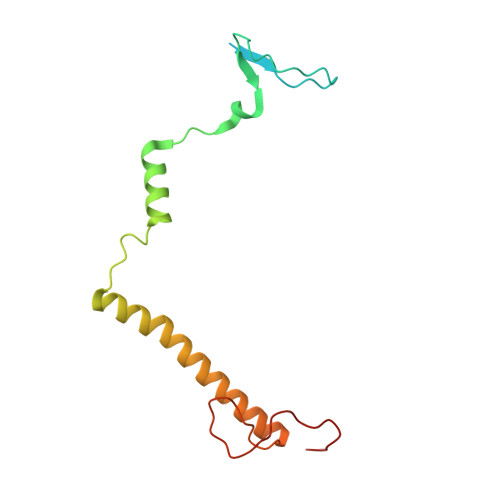
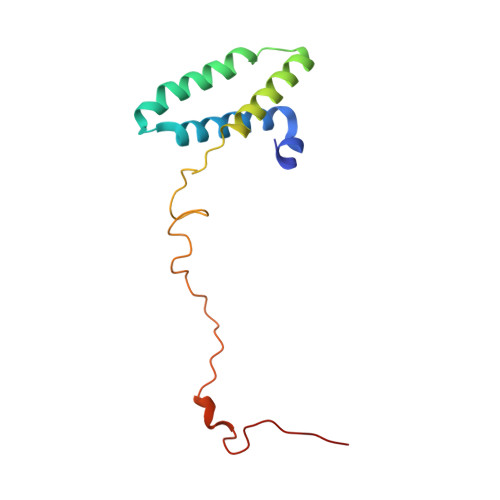
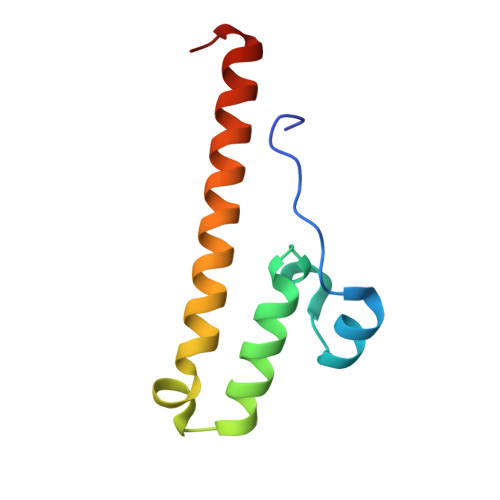
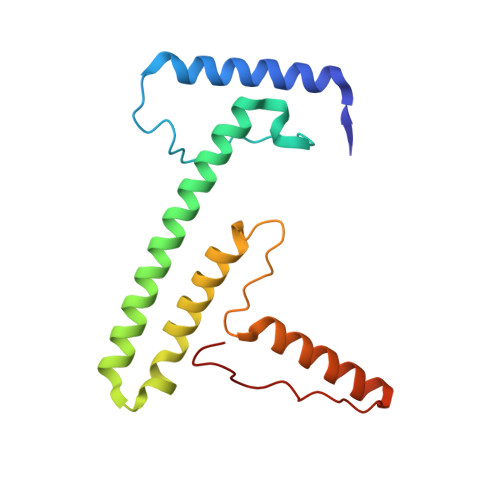
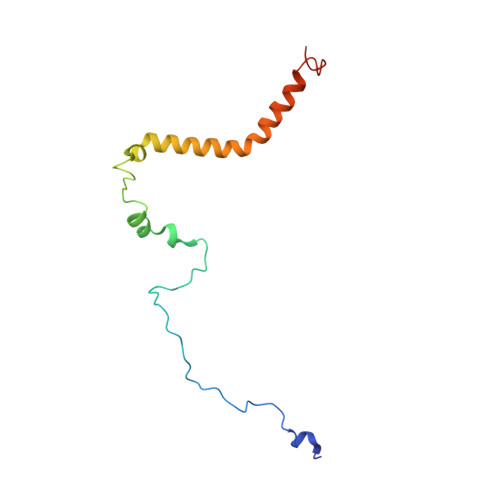
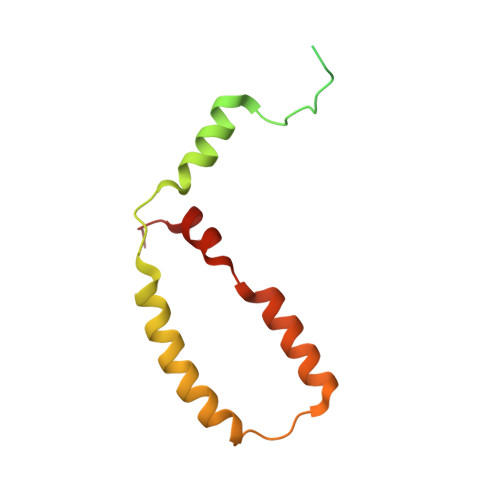
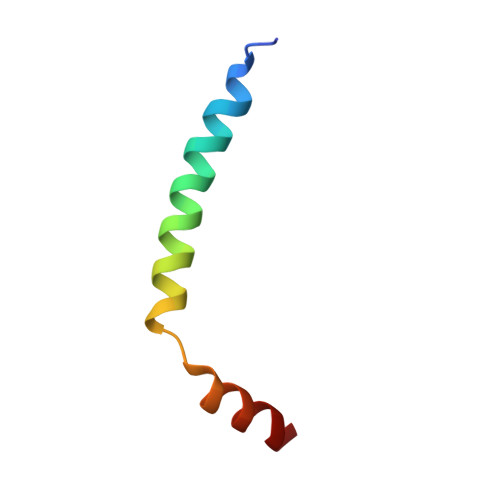
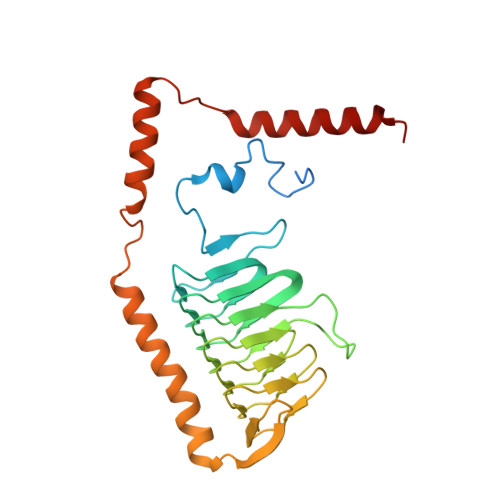
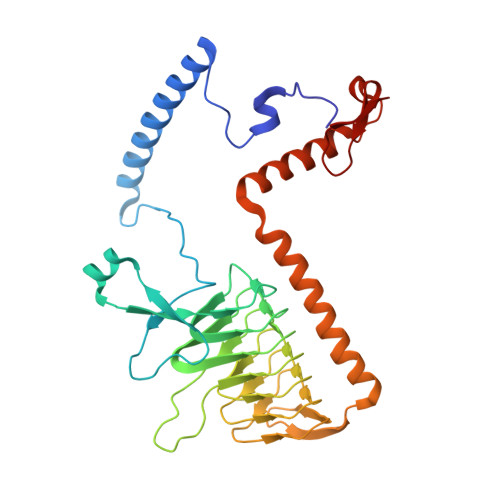
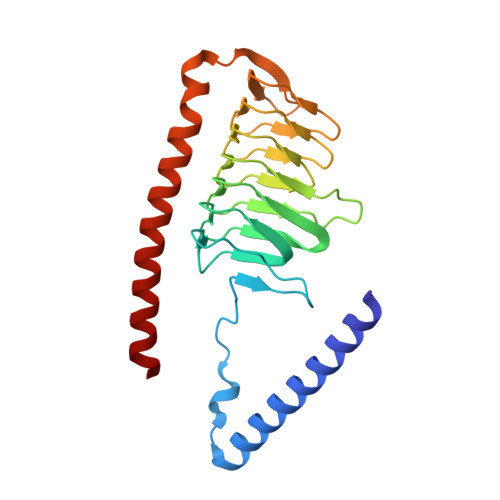
A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Klusch, N., Senkler, J., Yildiz, O., Kuhlbrandt, W., Braun, H.P.(2021) Plant Cell 33: 2072-2091
- PubMed: 33768254
- DOI: https://doi.org/10.1093/plcell/koab092
- Primary Citation of Related Structures:
7AQQ, 7AQR, 7AQW, 7AR7, 7AR8, 7AR9, 7ARB, 7ARC, 7ARD - PubMed Abstract:
Mitochondrial complex I is the main site for electron transfer to the respiratory chain and generates much of the proton gradient across the inner mitochondrial membrane. Complex I is composed of two arms, which form a conserved L-shape. We report the structures of the intact, 47-subunit mitochondrial complex I from Arabidopsis thaliana and the 51-subunit complex I from the green alga Polytomella sp., both at around 2.9 Å resolution. In both complexes, a heterotrimeric γ-carbonic anhydrase domain is attached to the membrane arm on the matrix side. Two states are resolved in A. thaliana complex I, with different angles between the two arms and different conformations of the ND1 (NADH dehydrogenase subunit 1) loop near the quinol binding site. The angle appears to depend on a bridge domain, which links the peripheral arm to the membrane arm and includes an unusual ferredoxin. We propose that the bridge domain participates in regulating the activity of plant complex I.
- Department of Structural Biology, Max-Planck-Institute of Biophysics, Frankfurt 60438, Germany.
Organizational Affiliation: