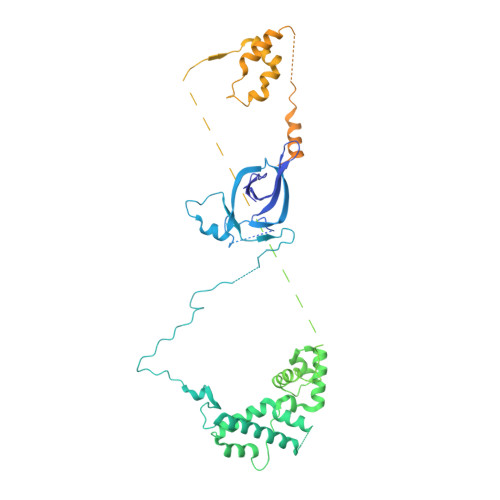
The topology of chromatin-binding domains in the NuRD deacetylase complex.
Millard, C.J., Fairall, L., Ragan, T.J., Savva, C.G., Schwabe, J.W.R.(2020) Nucleic Acids Res 48: 12972-12982
- PubMed: 33264408
- DOI: https://doi.org/10.1093/nar/gkaa1121
- Primary Citation of Related Structures:
7AO8, 7AO9, 7AOA - PubMed Abstract:
Class I histone deacetylase complexes play essential roles in many nuclear processes. Whilst they contain a common catalytic subunit, they have diverse modes of action determined by associated factors in the distinct complexes. The deacetylase module from the NuRD complex contains three protein domains that control the recruitment of chromatin to the deacetylase enzyme, HDAC1/2. Using biochemical approaches and cryo-electron microscopy, we have determined how three chromatin-binding domains (MTA1-BAH, MBD2/3 and RBBP4/7) are assembled in relation to the core complex so as to facilitate interaction of the complex with the genome. We observe a striking arrangement of the BAH domains suggesting a potential mechanism for binding to di-nucleosomes. We also find that the WD40 domains from RBBP4 are linked to the core with surprising flexibility that is likely important for chromatin engagement. A single MBD2 protein binds asymmetrically to the dimerisation interface of the complex. This symmetry mismatch explains the stoichiometry of the complex. Finally, our structures suggest how the holo-NuRD might assemble on a di-nucleosome substrate.
- The Leicester Institute of Structural and Chemical Biology, Department of Molecular and Cell Biology, University of Leicester, Leicester LE1 7RH, UK.
Organizational Affiliation: