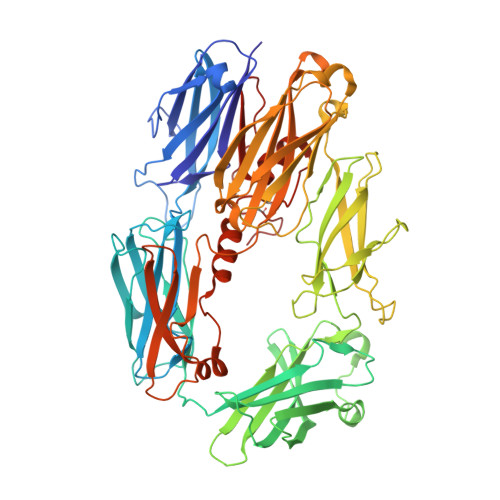
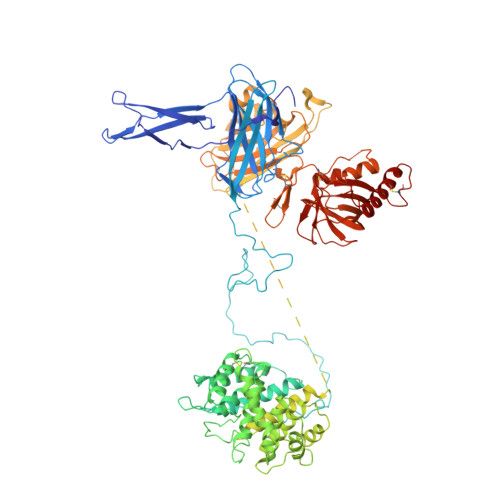
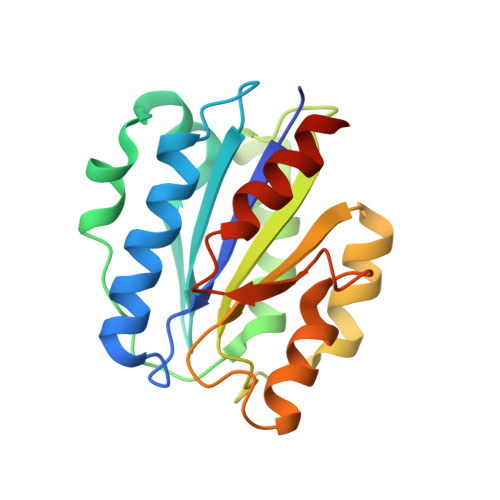
Entry History & Funding Information
Deposition Data
- Released Date: 2022-04-13
Deposition Author(s): Fernandez, F.J., Santos-Lopez, J., Martinez-Barricarte, R., Querol-Garcia, J., Navas-Yuste, S., Savko, M., Shepard, W.E., Rodriguez de Cordoba, S., Vega, M.C.
| Funding Organization | Location | Grant Number |
|---|
| Spanish Ministry of Science, Innovation, and Universities | Spain | RTI2018-102242-B-I00 |
| Spanish Ministry of Economy and Competitiveness | Spain | SAF2015-72961-EXP |
| Spanish Ministry of Economy and Competitiveness | Spain | SAF2015-66287-R |
| Other government | Spain | S2017/BMD-3673 |
| Spanish National Research Council | Spain | PIE-201620E064 |
- Version 1.0: 2022-04-13
Type: Initial release
- Version 1.1: 2022-04-20
Changes: Database references - Version 1.2: 2024-01-31
Changes: Data collection, Refinement description - Version 1.3: 2024-11-20
Changes: Structure summary