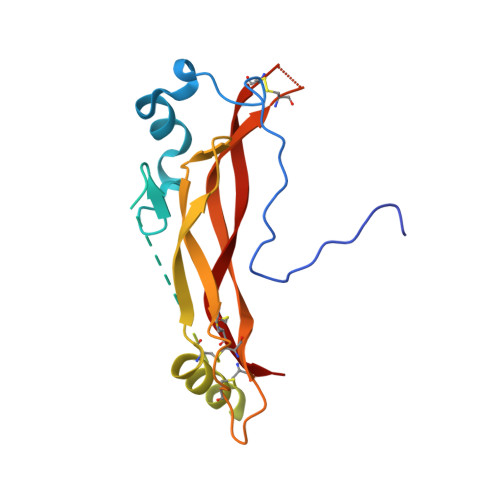
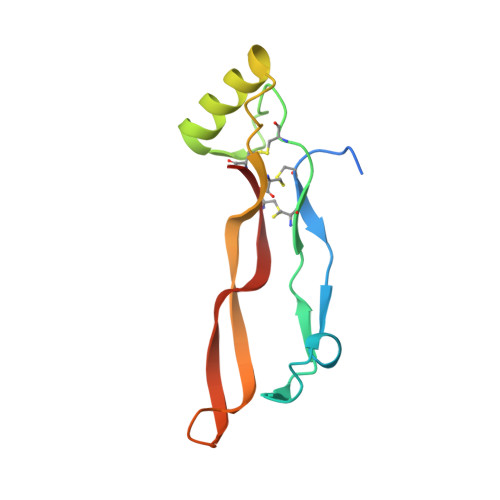
Structural analysis of the interaction between human cytokine BMP-2 and the antagonist Noggin reveals molecular details of cell chondrogenesis inhibition.
Robert, C., Kerff, F., Bouillenne, F., Gavage, M., Vandevenne, M., Filee, P., Matagne, A.(2023) J Biological Chem 299: 102892-102892
- PubMed: 36642181
- DOI: https://doi.org/10.1016/j.jbc.2023.102892
- Primary Citation of Related Structures:
7AG0 - PubMed Abstract:
Bone morphogenetic proteins (BMPs) are secreted cytokines belonging to the transforming growth factor-β superfamily. New therapeutic approaches based on BMP activity, particularly for cartilage and bone repair, have sparked considerable interest; however, a lack of understanding of their interaction pathways and the side effects associated with their use as biopharmaceuticals have dampened initial enthusiasm. Here, we used BMP-2 as a model system to gain further insight into both the relationship between structure and function in BMPs and the principles that govern affinity for their cognate antagonist Noggin. We produced BMP-2 and Noggin as inclusion bodies in Escherichia coli and developed simple and efficient protocols for preparing pure and homogeneous (in terms of size distribution) solutions of the native dimeric forms of the two proteins. The identity and integrity of the proteins were confirmed using mass spectrometry. Additionally, several in vitro cell-based assays, including enzymatic measurements, RT-qPCR, and matrix staining, demonstrated their biological activity during cell chondrogenic and hypertrophic differentiation. Furthermore, we characterized the simple 1:1 noncovalent interaction between the two ligands (K D ca. 0.4 nM) using bio-layer interferometry and solved the crystal structure of the complex using X-ray diffraction methods. We identified the residues and binding forces involved in the interaction between the two proteins. Finally, results obtained with the BMP-2 N102D mutant suggest that Noggin is remarkably flexible and able to accommodate major structural changes at the BMP-2 level. Altogether, our findings provide insights into BMP-2 activity and reveal the molecular details of its interaction with Noggin.
- Laboratory of Enzymology and Protein Folding, University of Liège, Liège, Belgium; Centre for Protein Engineering, InBioS Research Unit, University of Liège, Liège, Belgium.
Organizational Affiliation: