Domain mobility and allosteric activation of UbiD decarboxylases
Marshall, S.A., Payne, K.A.P., Leys, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phenolic acid decarboxylase | 480 | Sedimentibacter hydroxybenzoicus | Mutation(s): 0 Gene Names: shdC, ohb1 EC: 4.1.1.63 (PDB Primary Data), 4.1.1.61 (PDB Primary Data) | 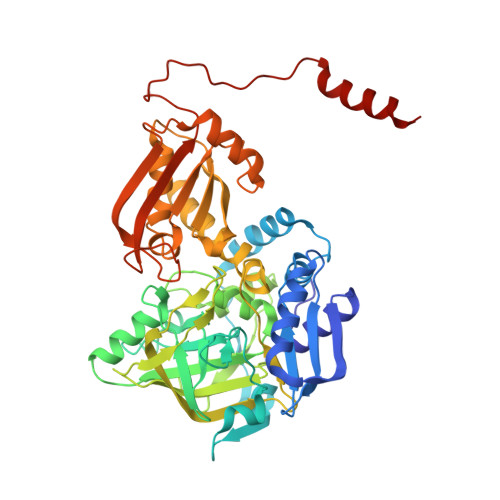 | |
UniProt | |||||
Find proteins for Q9S4M7 (Sedimentibacter hydroxybenzoicus) Explore Q9S4M7 Go to UniProtKB: Q9S4M7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9S4M7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein ShdD | 68 | Sedimentibacter hydroxybenzoicus | Mutation(s): 0 Gene Names: shdD | 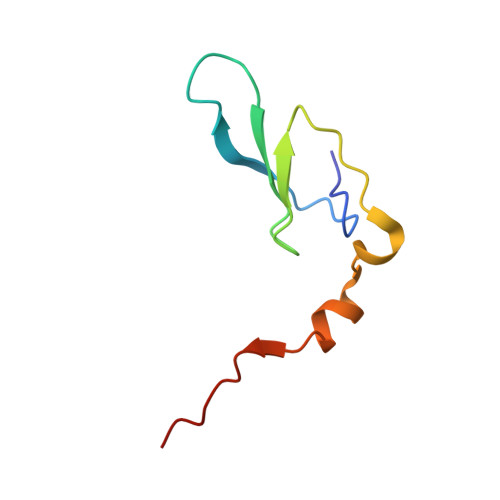 | |
UniProt | |||||
Find proteins for Q4R102 (Sedimentibacter hydroxybenzoicus) Explore Q4R102 Go to UniProtKB: Q4R102 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q4R102 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | AA [auth C] GA [auth D] M [auth A] NA [auth E] PA [auth E] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| PO4 Query on PO4 | DA [auth C] JA [auth D] Q [auth A] TA [auth E] X [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| ZN Query on ZN | AB [auth a] BB [auth b] CB [auth c] DB [auth d] EB [auth e] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | OA [auth E] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | FA [auth C] LA [auth D] MA [auth D] S [auth A] T [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | EA [auth C] KA [auth D] R [auth A] UA [auth E] Y [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | BA [auth C] CA [auth C] HA [auth D] IA [auth D] N [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 270.92 | α = 90 |
| b = 320.55 | β = 90 |
| c = 326.23 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | -- |