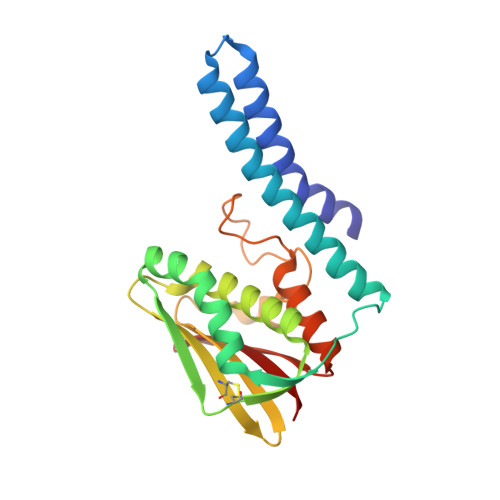
Crystal structure of the Escherichia coli CusS kinase core.
Cociurovscaia, A., Bujacz, G., Pietrzyk-Brzezinska, A.J.(2022) J Struct Biol 214: 107883-107883
- PubMed: 35907487
- DOI: https://doi.org/10.1016/j.jsb.2022.107883
- Primary Citation of Related Structures:
7ZP0 - PubMed Abstract:
The CusS histidine kinase is a member of Escherichia coli two-component signal transduction system, engaged in a response to copper ions excess in the cell periplasm. The periplasmic sensor domain of CusS binds the free copper ions and the CusS kinase core phosphorylates the cognate CusR which regulates transcription of the efflux pomp CusCBA. A small amount of copper ions is indispensable for the aerobic cell metabolism. Nonetheless, its excess in the cytoplasm generates damaging and reactive hydroxyl radicals. For that reason, understanding the bacterial copper sensing mechanisms can contribute to reducing bacterial copper-resistance and developing bactericidal copper-based materials. The crystal structure of the CusS kinase core was solved at the resolution of 1.4 Å. The cytoplasmic catalytic core domains formed a homodimer. Based on the obtained structure, the intramolecular and intermolecular interactions crucial for the mechanism of CusS autophosphorylation were described.
- Institute of Molecular and Industrial Biotechnology, Faculty of Biotechnology and Food Sciences, Lodz University of Technology, Lodz 90-537, Poland.
Organizational Affiliation: