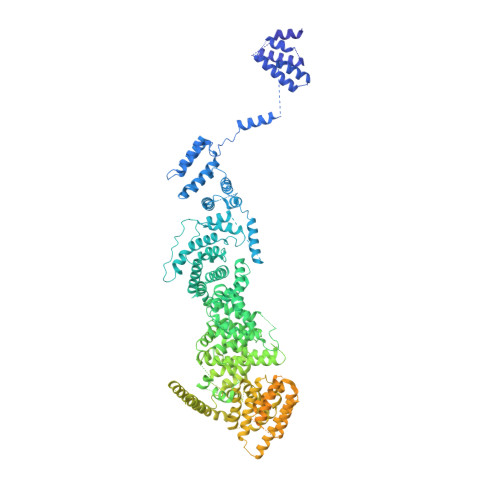
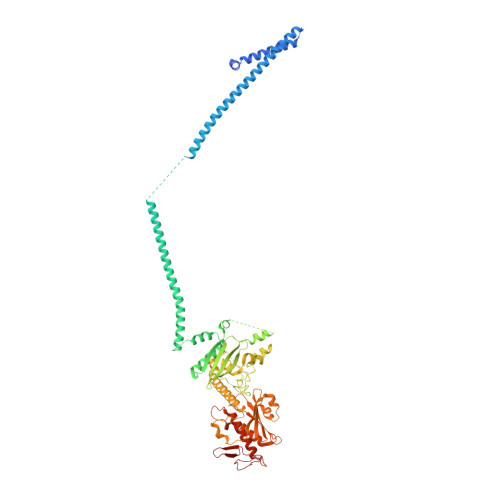
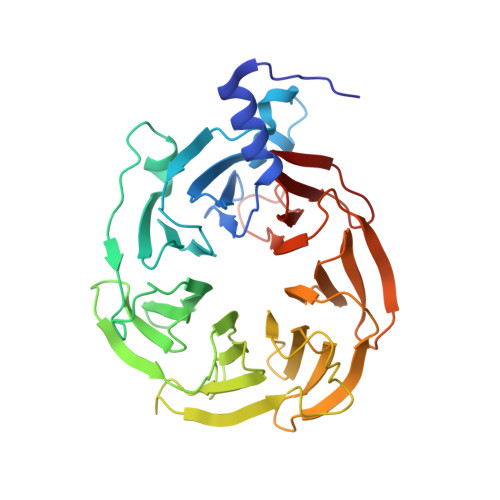
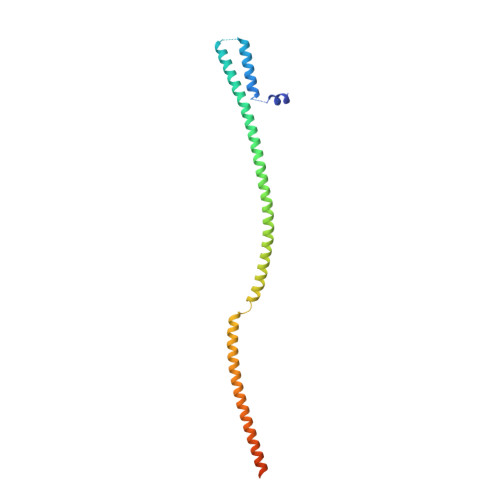
mRNA recognition and packaging by the human transcription-export complex.
Pacheco-Fiallos, B., Vorlander, M.K., Riabov-Bassat, D., Fin, L., O'Reilly, F.J., Ayala, F.I., Schellhaas, U., Rappsilber, J., Plaschka, C.(2023) Nature 616: 828-835
- PubMed: 37020021
- DOI: https://doi.org/10.1038/s41586-023-05904-0
- Primary Citation of Related Structures:
7ZNJ, 7ZNK, 7ZNL - PubMed Abstract:
Newly made mRNAs are processed and packaged into mature ribonucleoprotein complexes (mRNPs) and are recognized by the essential transcription-export complex (TREX) for nuclear export 1,2 . However, the mechanisms of mRNP recognition and three-dimensional mRNP organization are poorly understood 3 . Here we report cryo-electron microscopy and tomography structures of reconstituted and endogenous human mRNPs bound to the 2-MDa TREX complex. We show that mRNPs are recognized through multivalent interactions between the TREX subunit ALYREF and mRNP-bound exon junction complexes. Exon junction complexes can multimerize through ALYREF, which suggests a mechanism for mRNP organization. Endogenous mRNPs form compact globules that are coated by multiple TREX complexes. These results reveal how TREX may simultaneously recognize, compact and protect mRNAs to promote their packaging for nuclear export. The organization of mRNP globules provides a framework to understand how mRNP architecture facilitates mRNA biogenesis and export.
- Research Institute of Molecular Pathology (IMP), Vienna BioCenter (VBC), Vienna, Austria.
Organizational Affiliation: