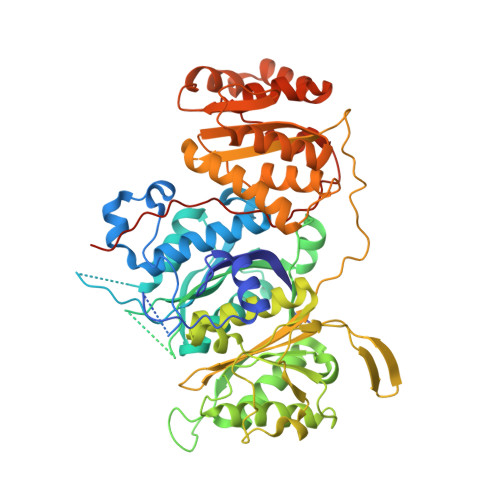
The structure of a polyketide synthase bimodule core.
Tittes, Y.U., Herbst, D.A., Martin, S.F.X., Munoz-Hernandez, H., Jakob, R.P., Maier, T.(2022) Sci Adv 8: eabo6918-eabo6918
- PubMed: 36129979
- DOI: https://doi.org/10.1126/sciadv.abo6918
- Primary Citation of Related Structures:
7ZM9, 7ZMA, 7ZMC, 7ZMD, 7ZMF, 7ZSK - PubMed Abstract:
Polyketide synthases (PKSs) are predominantly microbial biosynthetic enzymes. They assemble highly potent bioactive natural products from simple carboxylic acid precursors. The most versatile families of PKSs are organized as assembly lines of functional modules. Each module performs one round of precursor extension and optional modification, followed by directed transfer of the intermediate to the next module. While enzymatic domains and even modules of PKSs are well understood, the higher-order modular architecture of PKS assembly lines remains elusive. Here, we visualize a PKS bimodule core using cryo-electron microscopy and resolve a two-dimensional meshwork of the bimodule core formed by homotypic interactions between modules. The sheet-like organization provides the framework for efficient substrate transfer and for sequestration of trans-acting enzymes required for polyketide production.
- Biozentrum, University of Basel, Spitalstrasse 41, Basel 4056, Switzerland.
Organizational Affiliation: