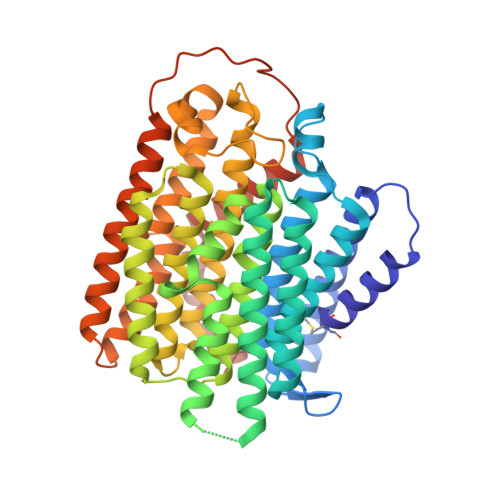
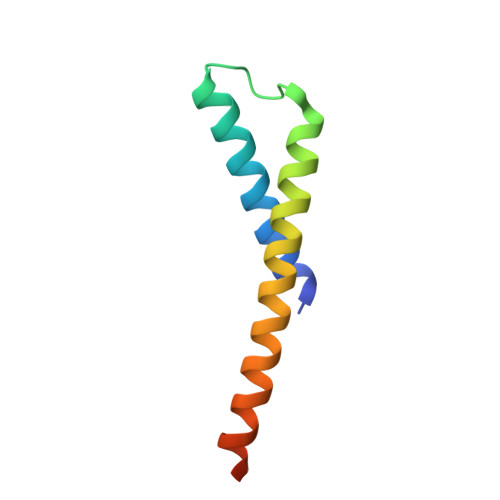
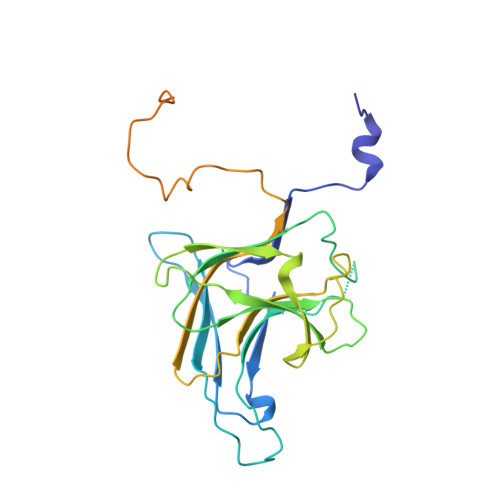
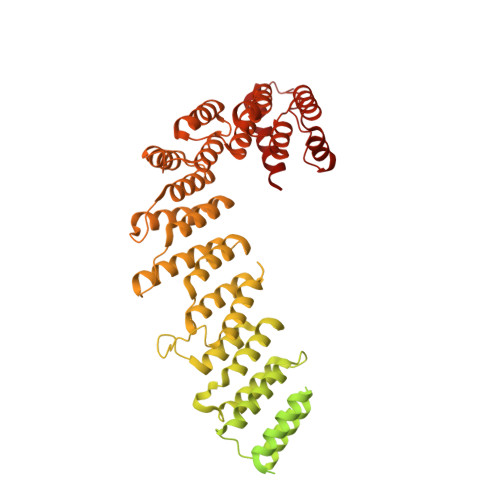
Insights into complex I assembly: Function of NDUFAF1 and a link with cardiolipin remodeling.
Schiller, J., Laube, E., Wittig, I., Kuhlbrandt, W., Vonck, J., Zickermann, V.(2022) Sci Adv 8: eadd3855-eadd3855
- PubMed: 36383672
- DOI: https://doi.org/10.1126/sciadv.add3855
- Primary Citation of Related Structures:
7ZKP, 7ZKQ - PubMed Abstract:
Respiratory complex I is a ~1-MDa proton pump in mitochondria. Its structure has been revealed in great detail, but the structural basis of its assembly, in humans involving at least 15 assembly factors, is essentially unknown. We determined cryo-electron microscopy structures of assembly intermediates associated with assembly factor NDUFAF1 in a yeast model system. Subunits ND2 and NDUFC2 together with assembly factors NDUFAF1 and CIA84 form the nucleation point of the NDUFAF1-dependent assembly pathway. Unexpectedly, the cardiolipin remodeling enzyme tafazzin is an integral component of this core complex. In a later intermediate, all 12 subunits of the proximal proton pump module have assembled. NDUFAF1 locks the central ND3 subunit in an assembly-competent conformation, and major rearrangements of central subunits are required for complex I maturation.
- Institute of Biochemistry II, University Hospital, Goethe University, 60590 Frankfurt am Main, Germany.
Organizational Affiliation: