Crystal Structure of Amycolatopsis jejuensis Multiple Inositol Polyphosphate Phosphatase, apo-protein
Acquistapace, I.M., Brearley, C.A., Hemmings, A.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Multiple inositol-polyphosphate phosphatase | 417 | Amycolatopsis jejuensis | Mutation(s): 0 EC: 3.1.3.62 (PDB Primary Data), 3.1.3.80 (UniProt) | 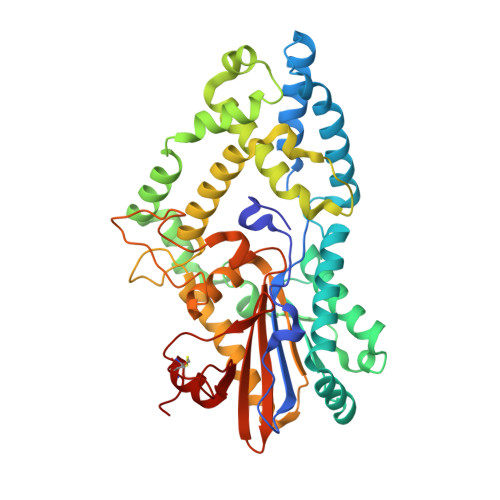 | |
UniProt | |||||
Find proteins for A0A915Q9M0 (Amycolatopsis jejuensis) Explore A0A915Q9M0 Go to UniProtKB: A0A915Q9M0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A915Q9M0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 68.848 | α = 90 |
| b = 87.137 | β = 97.402 |
| c = 70.358 | γ = 90 |
| Software Name | Purpose |
|---|---|
| GDA | data collection |
| PHENIX | refinement |
| xia2 | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/M022978/1 |