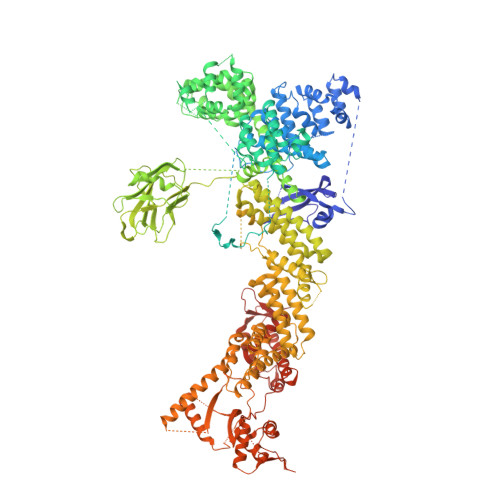
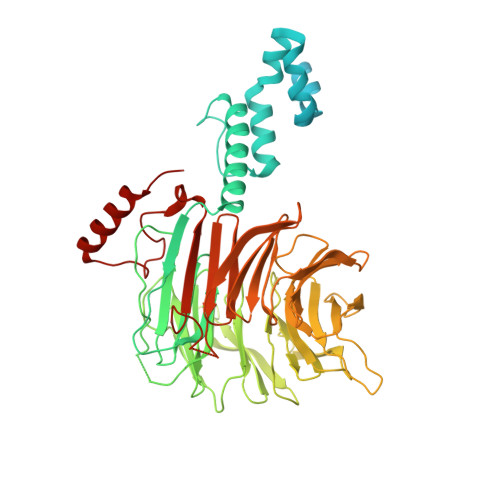
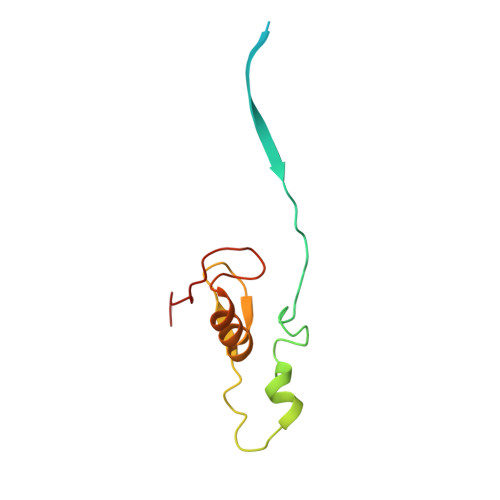
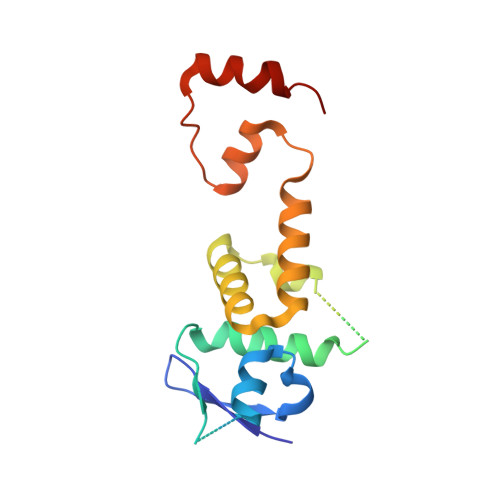
Structure of CRL7 FBXW8 reveals coupling with CUL1-RBX1/ROC1 for multi-cullin-RING E3-catalyzed ubiquitin ligation.
Hopf, L.V.M., Baek, K., Klugel, M., von Gronau, S., Xiong, Y., Schulman, B.A.(2022) Nat Struct Mol Biol 29: 854-862
- PubMed: 35982156
- DOI: https://doi.org/10.1038/s41594-022-00815-6
- Primary Citation of Related Structures:
7Z8B - PubMed Abstract:
Most cullin-RING ubiquitin ligases (CRLs) form homologous assemblies between a neddylated cullin-RING catalytic module and a variable substrate-binding receptor (for example, an F-box protein). However, the vertebrate-specific CRL7 FBXW8 is of interest because it eludes existing models, yet its constituent cullin CUL7 and F-box protein FBXW8 are essential for development, and CUL7 mutations cause 3M syndrome. In this study, cryo-EM and biochemical analyses reveal the CRL7 FBXW8 assembly. CUL7's exclusivity for FBXW8 among all F-box proteins is explained by its unique F-box-independent binding mode. In CRL7 FBXW8 , the RBX1 (also known as ROC1) RING domain is constrained in an orientation incompatible with binding E2~NEDD8 or E2~ubiquitin intermediates. Accordingly, purified recombinant CRL7 FBXW8 lacks auto-neddylation and ubiquitination activities. Instead, our data indicate that CRL7 serves as a substrate receptor linked via SKP1-FBXW8 to a neddylated CUL1-RBX1 catalytic module mediating ubiquitination. The structure reveals a distinctive CRL-CRL partnership, and provides a framework for understanding CUL7 assemblies safeguarding human health.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: